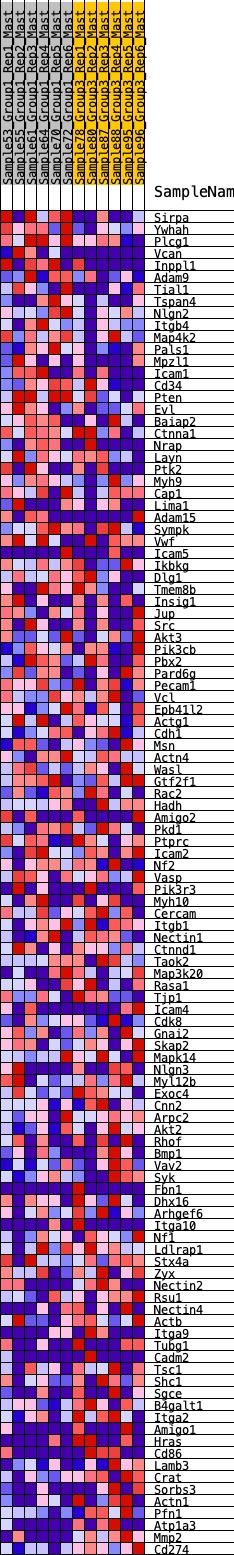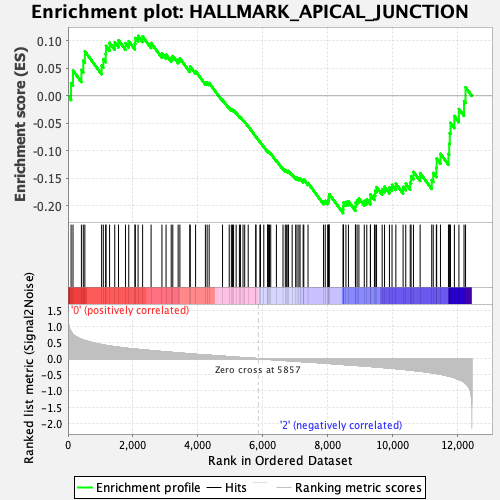
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | -0.21349211 |
| Normalized Enrichment Score (NES) | -0.9587465 |
| Nominal p-value | 0.5304878 |
| FDR q-value | 0.9145504 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sirpa | 95 | 0.831 | 0.0233 | No |
| 2 | Ywhah | 154 | 0.737 | 0.0461 | No |
| 3 | Plcg1 | 413 | 0.595 | 0.0474 | No |
| 4 | Vcan | 471 | 0.573 | 0.0641 | No |
| 5 | Inppl1 | 519 | 0.560 | 0.0812 | No |
| 6 | Adam9 | 1036 | 0.431 | 0.0554 | No |
| 7 | Tial1 | 1088 | 0.420 | 0.0670 | No |
| 8 | Tspan4 | 1161 | 0.411 | 0.0765 | No |
| 9 | Nlgn2 | 1171 | 0.408 | 0.0910 | No |
| 10 | Itgb4 | 1281 | 0.390 | 0.0967 | No |
| 11 | Map4k2 | 1440 | 0.367 | 0.0976 | No |
| 12 | Pals1 | 1556 | 0.351 | 0.1013 | No |
| 13 | Mpzl1 | 1771 | 0.325 | 0.0961 | No |
| 14 | Icam1 | 1872 | 0.313 | 0.0997 | No |
| 15 | Cd34 | 2061 | 0.295 | 0.0954 | No |
| 16 | Pten | 2074 | 0.294 | 0.1054 | No |
| 17 | Evl | 2159 | 0.285 | 0.1092 | No |
| 18 | Baiap2 | 2299 | 0.272 | 0.1081 | No |
| 19 | Ctnna1 | 2560 | 0.249 | 0.0963 | No |
| 20 | Nrap | 2894 | 0.218 | 0.0775 | No |
| 21 | Layn | 3021 | 0.208 | 0.0750 | No |
| 22 | Ptk2 | 3180 | 0.194 | 0.0694 | No |
| 23 | Myh9 | 3229 | 0.190 | 0.0726 | No |
| 24 | Cap1 | 3393 | 0.177 | 0.0660 | No |
| 25 | Lima1 | 3447 | 0.172 | 0.0682 | No |
| 26 | Adam15 | 3752 | 0.148 | 0.0490 | No |
| 27 | Sympk | 3765 | 0.147 | 0.0535 | No |
| 28 | Vwf | 3931 | 0.132 | 0.0451 | No |
| 29 | Icam5 | 4233 | 0.112 | 0.0249 | No |
| 30 | Ikbkg | 4287 | 0.108 | 0.0246 | No |
| 31 | Dlg1 | 4349 | 0.104 | 0.0236 | No |
| 32 | Tmem8b | 4761 | 0.074 | -0.0070 | No |
| 33 | Insig1 | 4967 | 0.058 | -0.0215 | No |
| 34 | Jup | 5033 | 0.055 | -0.0247 | No |
| 35 | Src | 5067 | 0.052 | -0.0254 | No |
| 36 | Akt3 | 5101 | 0.050 | -0.0262 | No |
| 37 | Pik3cb | 5180 | 0.044 | -0.0309 | No |
| 38 | Pbx2 | 5281 | 0.036 | -0.0377 | No |
| 39 | Pard6g | 5313 | 0.034 | -0.0389 | No |
| 40 | Pecam1 | 5392 | 0.029 | -0.0442 | No |
| 41 | Vcl | 5444 | 0.025 | -0.0474 | No |
| 42 | Epb41l2 | 5552 | 0.018 | -0.0554 | No |
| 43 | Actg1 | 5784 | 0.004 | -0.0739 | No |
| 44 | Cdh1 | 5791 | 0.004 | -0.0743 | No |
| 45 | Msn | 5912 | -0.003 | -0.0839 | No |
| 46 | Actn4 | 5931 | -0.004 | -0.0852 | No |
| 47 | Wasl | 6032 | -0.014 | -0.0928 | No |
| 48 | Gtf2f1 | 6151 | -0.022 | -0.1015 | No |
| 49 | Rac2 | 6154 | -0.022 | -0.1009 | No |
| 50 | Hadh | 6180 | -0.024 | -0.1020 | No |
| 51 | Amigo2 | 6208 | -0.025 | -0.1033 | No |
| 52 | Pkd1 | 6246 | -0.028 | -0.1052 | No |
| 53 | Ptprc | 6421 | -0.039 | -0.1179 | No |
| 54 | Icam2 | 6624 | -0.053 | -0.1323 | No |
| 55 | Nf2 | 6695 | -0.058 | -0.1358 | No |
| 56 | Vasp | 6727 | -0.060 | -0.1361 | No |
| 57 | Pik3r3 | 6773 | -0.062 | -0.1374 | No |
| 58 | Myh10 | 6795 | -0.064 | -0.1367 | No |
| 59 | Cercam | 6906 | -0.071 | -0.1430 | No |
| 60 | Itgb1 | 7014 | -0.078 | -0.1488 | No |
| 61 | Nectin1 | 7056 | -0.082 | -0.1491 | No |
| 62 | Ctnnd1 | 7114 | -0.086 | -0.1505 | No |
| 63 | Taok2 | 7150 | -0.089 | -0.1500 | No |
| 64 | Map3k20 | 7245 | -0.095 | -0.1540 | No |
| 65 | Rasa1 | 7265 | -0.096 | -0.1520 | No |
| 66 | Tjp1 | 7395 | -0.103 | -0.1586 | No |
| 67 | Icam4 | 7873 | -0.136 | -0.1922 | No |
| 68 | Cdk8 | 7925 | -0.140 | -0.1911 | No |
| 69 | Gnai2 | 8000 | -0.145 | -0.1917 | No |
| 70 | Skap2 | 8026 | -0.147 | -0.1883 | No |
| 71 | Mapk14 | 8038 | -0.148 | -0.1837 | No |
| 72 | Nlgn3 | 8048 | -0.148 | -0.1789 | No |
| 73 | Myl12b | 8476 | -0.181 | -0.2067 | Yes |
| 74 | Exoc4 | 8482 | -0.181 | -0.2004 | Yes |
| 75 | Cnn2 | 8489 | -0.182 | -0.1941 | Yes |
| 76 | Arpc2 | 8563 | -0.188 | -0.1930 | Yes |
| 77 | Akt2 | 8640 | -0.193 | -0.1919 | Yes |
| 78 | Rhof | 8855 | -0.207 | -0.2015 | Yes |
| 79 | Bmp1 | 8860 | -0.208 | -0.1941 | Yes |
| 80 | Vav2 | 8907 | -0.211 | -0.1900 | Yes |
| 81 | Syk | 8963 | -0.215 | -0.1864 | Yes |
| 82 | Fbn1 | 9128 | -0.227 | -0.1912 | Yes |
| 83 | Dhx16 | 9205 | -0.234 | -0.1886 | Yes |
| 84 | Arhgef6 | 9317 | -0.243 | -0.1886 | Yes |
| 85 | Itga10 | 9318 | -0.243 | -0.1795 | Yes |
| 86 | Nf1 | 9441 | -0.252 | -0.1800 | Yes |
| 87 | Ldlrap1 | 9464 | -0.254 | -0.1723 | Yes |
| 88 | Stx4a | 9506 | -0.258 | -0.1660 | Yes |
| 89 | Zyx | 9679 | -0.271 | -0.1699 | Yes |
| 90 | Nectin2 | 9752 | -0.278 | -0.1653 | Yes |
| 91 | Rsu1 | 9903 | -0.292 | -0.1666 | Yes |
| 92 | Nectin4 | 9983 | -0.298 | -0.1619 | Yes |
| 93 | Actb | 10101 | -0.308 | -0.1599 | Yes |
| 94 | Itga9 | 10324 | -0.330 | -0.1656 | Yes |
| 95 | Tubg1 | 10406 | -0.338 | -0.1595 | Yes |
| 96 | Cadm2 | 10543 | -0.354 | -0.1574 | Yes |
| 97 | Tsc1 | 10570 | -0.356 | -0.1462 | Yes |
| 98 | Shc1 | 10643 | -0.365 | -0.1384 | Yes |
| 99 | Sgce | 10849 | -0.387 | -0.1406 | Yes |
| 100 | B4galt1 | 11206 | -0.438 | -0.1531 | Yes |
| 101 | Itga2 | 11253 | -0.445 | -0.1402 | Yes |
| 102 | Amigo1 | 11348 | -0.462 | -0.1306 | Yes |
| 103 | Hras | 11359 | -0.463 | -0.1142 | Yes |
| 104 | Cd86 | 11475 | -0.481 | -0.1055 | Yes |
| 105 | Lamb3 | 11722 | -0.537 | -0.1054 | Yes |
| 106 | Crat | 11745 | -0.544 | -0.0869 | Yes |
| 107 | Sorbs3 | 11762 | -0.546 | -0.0679 | Yes |
| 108 | Actn1 | 11785 | -0.551 | -0.0491 | Yes |
| 109 | Pfn1 | 11904 | -0.590 | -0.0366 | Yes |
| 110 | Atp1a3 | 12044 | -0.644 | -0.0239 | Yes |
| 111 | Mmp2 | 12201 | -0.729 | -0.0093 | Yes |
| 112 | Cd274 | 12248 | -0.770 | 0.0157 | Yes |