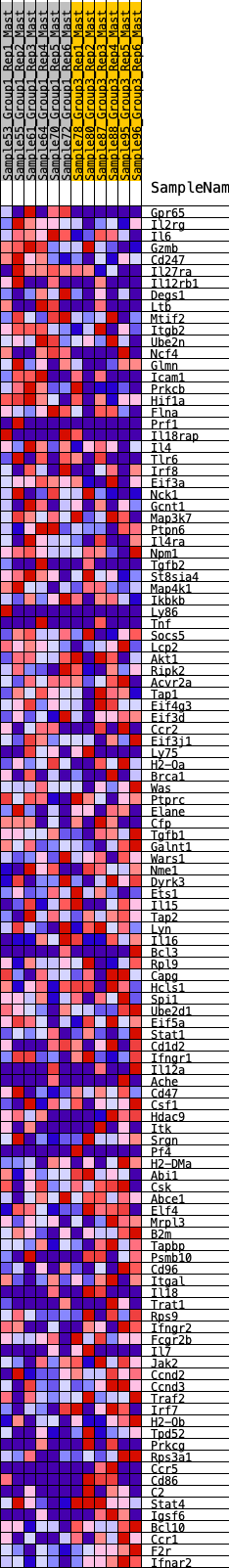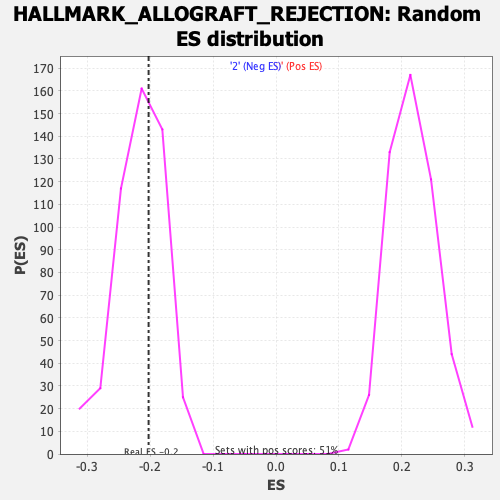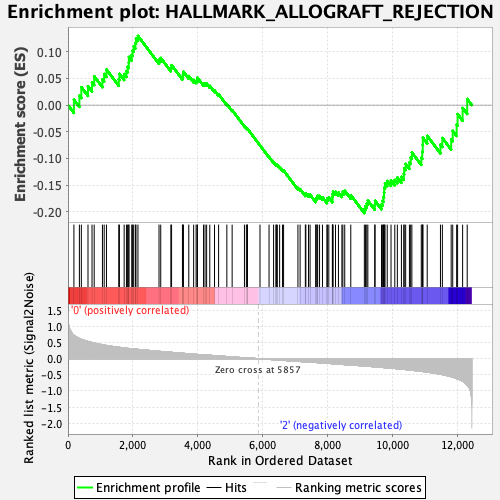
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group3.Mast_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_ALLOGRAFT_REJECTION |
| Enrichment Score (ES) | -0.2025927 |
| Normalized Enrichment Score (NES) | -0.9341109 |
| Nominal p-value | 0.620202 |
| FDR q-value | 0.853497 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gpr65 | 182 | 0.720 | 0.0101 | No |
| 2 | Il2rg | 351 | 0.619 | 0.0179 | No |
| 3 | Il6 | 411 | 0.596 | 0.0337 | No |
| 4 | Gzmb | 616 | 0.531 | 0.0355 | No |
| 5 | Cd247 | 740 | 0.499 | 0.0428 | No |
| 6 | Il27ra | 807 | 0.481 | 0.0541 | No |
| 7 | Il12rb1 | 1061 | 0.426 | 0.0483 | No |
| 8 | Degs1 | 1115 | 0.417 | 0.0584 | No |
| 9 | Ltb | 1185 | 0.406 | 0.0668 | No |
| 10 | Mtif2 | 1564 | 0.349 | 0.0482 | No |
| 11 | Itgb2 | 1584 | 0.348 | 0.0587 | No |
| 12 | Ube2n | 1730 | 0.330 | 0.0583 | No |
| 13 | Ncf4 | 1801 | 0.321 | 0.0638 | No |
| 14 | Glmn | 1836 | 0.316 | 0.0719 | No |
| 15 | Icam1 | 1872 | 0.313 | 0.0799 | No |
| 16 | Prkcb | 1877 | 0.313 | 0.0904 | No |
| 17 | Hif1a | 1964 | 0.305 | 0.0940 | No |
| 18 | Flna | 1990 | 0.302 | 0.1024 | No |
| 19 | Prf1 | 2023 | 0.299 | 0.1101 | No |
| 20 | Il18rap | 2076 | 0.294 | 0.1161 | No |
| 21 | Il4 | 2094 | 0.292 | 0.1248 | No |
| 22 | Tlr6 | 2155 | 0.285 | 0.1298 | No |
| 23 | Irf8 | 2804 | 0.227 | 0.0851 | No |
| 24 | Eif3a | 2857 | 0.221 | 0.0885 | No |
| 25 | Nck1 | 3169 | 0.196 | 0.0700 | No |
| 26 | Gcnt1 | 3191 | 0.193 | 0.0750 | No |
| 27 | Map3k7 | 3526 | 0.165 | 0.0536 | No |
| 28 | Ptpn6 | 3543 | 0.164 | 0.0580 | No |
| 29 | Il4ra | 3555 | 0.163 | 0.0627 | No |
| 30 | Npm1 | 3721 | 0.151 | 0.0546 | No |
| 31 | Tgfb2 | 3871 | 0.138 | 0.0472 | No |
| 32 | St8sia4 | 3947 | 0.131 | 0.0457 | No |
| 33 | Map4k1 | 3982 | 0.128 | 0.0474 | No |
| 34 | Ikbkb | 3986 | 0.128 | 0.0516 | No |
| 35 | Ly86 | 4176 | 0.114 | 0.0402 | No |
| 36 | Tnf | 4222 | 0.113 | 0.0404 | No |
| 37 | Socs5 | 4271 | 0.109 | 0.0403 | No |
| 38 | Lcp2 | 4368 | 0.103 | 0.0361 | No |
| 39 | Akt1 | 4514 | 0.092 | 0.0275 | No |
| 40 | Ripk2 | 4640 | 0.083 | 0.0202 | No |
| 41 | Acvr2a | 4894 | 0.064 | 0.0019 | No |
| 42 | Tap1 | 5057 | 0.053 | -0.0094 | No |
| 43 | Eif4g3 | 5436 | 0.026 | -0.0392 | No |
| 44 | Eif3d | 5495 | 0.022 | -0.0431 | No |
| 45 | Ccr2 | 5522 | 0.020 | -0.0445 | No |
| 46 | Eif3j1 | 5528 | 0.020 | -0.0443 | No |
| 47 | Ly75 | 5915 | -0.003 | -0.0755 | No |
| 48 | H2-Oa | 6194 | -0.024 | -0.0972 | No |
| 49 | Brca1 | 6334 | -0.033 | -0.1073 | No |
| 50 | Was | 6405 | -0.038 | -0.1117 | No |
| 51 | Ptprc | 6421 | -0.039 | -0.1115 | No |
| 52 | Elane | 6449 | -0.041 | -0.1123 | No |
| 53 | Cfp | 6520 | -0.046 | -0.1164 | No |
| 54 | Tgfb1 | 6608 | -0.052 | -0.1217 | No |
| 55 | Galnt1 | 6642 | -0.054 | -0.1225 | No |
| 56 | Wars1 | 7084 | -0.084 | -0.1554 | No |
| 57 | Nme1 | 7151 | -0.089 | -0.1576 | No |
| 58 | Dyrk3 | 7312 | -0.099 | -0.1672 | No |
| 59 | Ets1 | 7328 | -0.099 | -0.1650 | No |
| 60 | Il15 | 7410 | -0.104 | -0.1680 | No |
| 61 | Tap2 | 7458 | -0.107 | -0.1681 | No |
| 62 | Lyn | 7631 | -0.119 | -0.1779 | No |
| 63 | Il16 | 7644 | -0.119 | -0.1748 | No |
| 64 | Bcl3 | 7672 | -0.122 | -0.1728 | No |
| 65 | Rpl9 | 7692 | -0.123 | -0.1700 | No |
| 66 | Capg | 7750 | -0.127 | -0.1703 | No |
| 67 | Hcls1 | 7840 | -0.133 | -0.1729 | No |
| 68 | Spi1 | 7978 | -0.143 | -0.1790 | No |
| 69 | Ube2d1 | 7989 | -0.144 | -0.1749 | No |
| 70 | Eif5a | 8035 | -0.147 | -0.1734 | No |
| 71 | Stat1 | 8146 | -0.156 | -0.1770 | No |
| 72 | Cd1d2 | 8147 | -0.156 | -0.1716 | No |
| 73 | Ifngr1 | 8154 | -0.156 | -0.1666 | No |
| 74 | Il12a | 8166 | -0.157 | -0.1621 | No |
| 75 | Ache | 8242 | -0.163 | -0.1625 | No |
| 76 | Cd47 | 8332 | -0.170 | -0.1639 | No |
| 77 | Csf1 | 8439 | -0.178 | -0.1663 | No |
| 78 | Hdac9 | 8468 | -0.180 | -0.1624 | No |
| 79 | Itk | 8526 | -0.185 | -0.1606 | No |
| 80 | Srgn | 8712 | -0.198 | -0.1688 | No |
| 81 | Pf4 | 9130 | -0.227 | -0.1947 | Yes |
| 82 | H2-DMa | 9164 | -0.231 | -0.1894 | Yes |
| 83 | Abi1 | 9204 | -0.234 | -0.1845 | Yes |
| 84 | Csk | 9238 | -0.236 | -0.1790 | Yes |
| 85 | Abce1 | 9451 | -0.253 | -0.1875 | Yes |
| 86 | Elf4 | 9460 | -0.253 | -0.1794 | Yes |
| 87 | Mrpl3 | 9659 | -0.269 | -0.1861 | Yes |
| 88 | B2m | 9693 | -0.273 | -0.1794 | Yes |
| 89 | Tapbp | 9720 | -0.276 | -0.1719 | Yes |
| 90 | Psmb10 | 9736 | -0.277 | -0.1636 | Yes |
| 91 | Cd96 | 9744 | -0.277 | -0.1546 | Yes |
| 92 | Itgal | 9767 | -0.279 | -0.1467 | Yes |
| 93 | Il18 | 9835 | -0.285 | -0.1423 | Yes |
| 94 | Trat1 | 9952 | -0.296 | -0.1414 | Yes |
| 95 | Rps9 | 10068 | -0.306 | -0.1402 | Yes |
| 96 | Ifngr2 | 10147 | -0.313 | -0.1357 | Yes |
| 97 | Fcgr2b | 10272 | -0.326 | -0.1345 | Yes |
| 98 | Il7 | 10346 | -0.332 | -0.1290 | Yes |
| 99 | Jak2 | 10357 | -0.333 | -0.1183 | Yes |
| 100 | Ccnd2 | 10403 | -0.338 | -0.1102 | Yes |
| 101 | Ccnd3 | 10519 | -0.350 | -0.1075 | Yes |
| 102 | Traf2 | 10557 | -0.354 | -0.0982 | Yes |
| 103 | Irf7 | 10594 | -0.359 | -0.0887 | Yes |
| 104 | H2-Ob | 10884 | -0.391 | -0.0986 | Yes |
| 105 | Tpd52 | 10914 | -0.396 | -0.0873 | Yes |
| 106 | Prkcg | 10929 | -0.398 | -0.0747 | Yes |
| 107 | Rps3a1 | 10933 | -0.399 | -0.0611 | Yes |
| 108 | Ccr5 | 11067 | -0.415 | -0.0576 | Yes |
| 109 | Cd86 | 11475 | -0.481 | -0.0740 | Yes |
| 110 | C2 | 11534 | -0.494 | -0.0616 | Yes |
| 111 | Stat4 | 11805 | -0.558 | -0.0642 | Yes |
| 112 | Igsf6 | 11853 | -0.573 | -0.0482 | Yes |
| 113 | Bcl10 | 11974 | -0.621 | -0.0365 | Yes |
| 114 | Ccr1 | 12003 | -0.632 | -0.0169 | Yes |
| 115 | F2r | 12158 | -0.693 | -0.0054 | Yes |
| 116 | Ifnar2 | 12299 | -0.818 | 0.0115 | Yes |