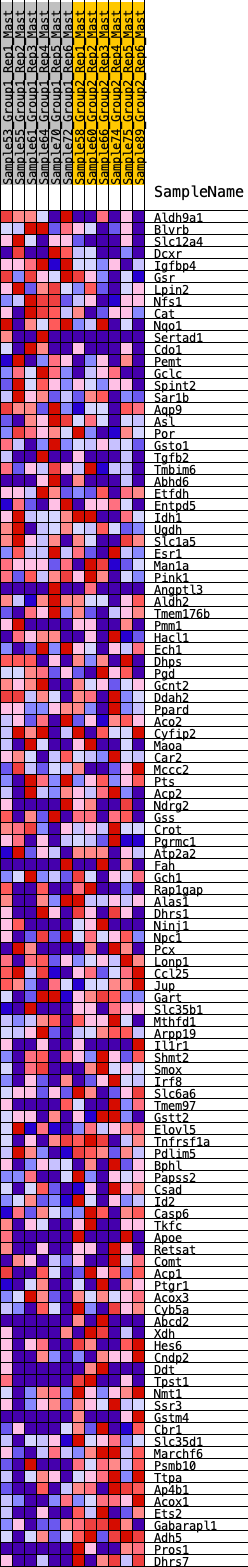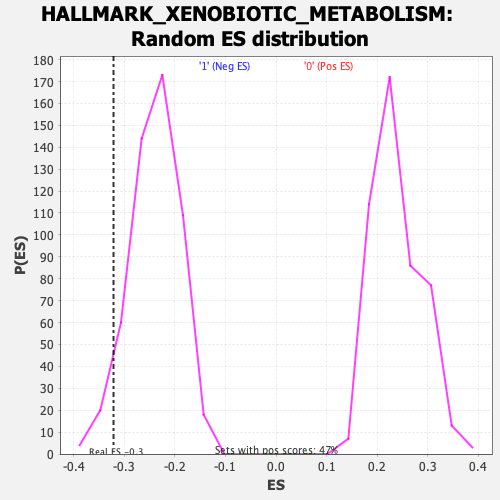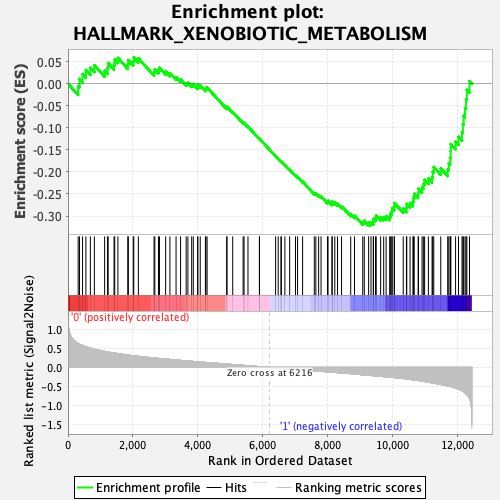
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.32167098 |
| Normalized Enrichment Score (NES) | -1.3433951 |
| Nominal p-value | 0.056818184 |
| FDR q-value | 0.30933544 |
| FWER p-Value | 0.666 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Aldh9a1 | 313 | 0.630 | -0.0053 | No |
| 2 | Blvrb | 355 | 0.611 | 0.0109 | No |
| 3 | Slc12a4 | 448 | 0.578 | 0.0219 | No |
| 4 | Dcxr | 551 | 0.547 | 0.0312 | No |
| 5 | Igfbp4 | 689 | 0.510 | 0.0363 | No |
| 6 | Gsr | 812 | 0.480 | 0.0418 | No |
| 7 | Lpin2 | 1129 | 0.423 | 0.0297 | No |
| 8 | Nfs1 | 1220 | 0.406 | 0.0353 | No |
| 9 | Cat | 1236 | 0.404 | 0.0470 | No |
| 10 | Nqo1 | 1418 | 0.380 | 0.0445 | No |
| 11 | Sertad1 | 1439 | 0.377 | 0.0549 | No |
| 12 | Cdo1 | 1538 | 0.363 | 0.0586 | No |
| 13 | Pemt | 1842 | 0.326 | 0.0444 | No |
| 14 | Gclc | 1859 | 0.324 | 0.0535 | No |
| 15 | Spint2 | 2009 | 0.308 | 0.0512 | No |
| 16 | Sar1b | 2026 | 0.306 | 0.0597 | No |
| 17 | Aqp9 | 2168 | 0.292 | 0.0576 | No |
| 18 | Asl | 2651 | 0.250 | 0.0265 | No |
| 19 | Por | 2675 | 0.247 | 0.0325 | No |
| 20 | Gsto1 | 2787 | 0.238 | 0.0311 | No |
| 21 | Tgfb2 | 2818 | 0.235 | 0.0362 | No |
| 22 | Tmbim6 | 3008 | 0.220 | 0.0279 | No |
| 23 | Abhd6 | 3140 | 0.210 | 0.0240 | No |
| 24 | Etfdh | 3329 | 0.195 | 0.0150 | No |
| 25 | Entpd5 | 3468 | 0.184 | 0.0097 | No |
| 26 | Idh1 | 3640 | 0.172 | 0.0013 | No |
| 27 | Ugdh | 3694 | 0.169 | 0.0024 | No |
| 28 | Slc1a5 | 3810 | 0.161 | -0.0017 | No |
| 29 | Esr1 | 3860 | 0.158 | -0.0007 | No |
| 30 | Man1a | 3997 | 0.147 | -0.0070 | No |
| 31 | Pink1 | 3998 | 0.147 | -0.0023 | No |
| 32 | Angptl3 | 4074 | 0.143 | -0.0038 | No |
| 33 | Aldh2 | 4233 | 0.130 | -0.0125 | No |
| 34 | Tmem176b | 4248 | 0.129 | -0.0095 | No |
| 35 | Pmm1 | 4285 | 0.127 | -0.0083 | No |
| 36 | Hacl1 | 4889 | 0.090 | -0.0544 | No |
| 37 | Ech1 | 4899 | 0.088 | -0.0523 | No |
| 38 | Dhps | 5075 | 0.075 | -0.0641 | No |
| 39 | Pgd | 5395 | 0.055 | -0.0882 | No |
| 40 | Gcnt2 | 5424 | 0.052 | -0.0888 | No |
| 41 | Ddah2 | 5545 | 0.044 | -0.0971 | No |
| 42 | Ppard | 5896 | 0.021 | -0.1249 | No |
| 43 | Aco2 | 5900 | 0.021 | -0.1244 | No |
| 44 | Cyfip2 | 6396 | -0.011 | -0.1642 | No |
| 45 | Maoa | 6476 | -0.016 | -0.1701 | No |
| 46 | Car2 | 6561 | -0.023 | -0.1762 | No |
| 47 | Mccc2 | 6574 | -0.023 | -0.1764 | No |
| 48 | Pts | 6682 | -0.029 | -0.1842 | No |
| 49 | Acp2 | 6829 | -0.037 | -0.1949 | No |
| 50 | Ndrg2 | 7008 | -0.049 | -0.2078 | No |
| 51 | Gss | 7073 | -0.053 | -0.2113 | No |
| 52 | Crot | 7228 | -0.063 | -0.2217 | No |
| 53 | Pgrmc1 | 7588 | -0.089 | -0.2480 | No |
| 54 | Atp2a2 | 7636 | -0.092 | -0.2489 | No |
| 55 | Fah | 7722 | -0.097 | -0.2527 | No |
| 56 | Gch1 | 7796 | -0.100 | -0.2554 | No |
| 57 | Rap1gap | 7994 | -0.115 | -0.2677 | No |
| 58 | Alas1 | 8012 | -0.117 | -0.2653 | No |
| 59 | Dhrs1 | 8135 | -0.125 | -0.2712 | No |
| 60 | Ninj1 | 8136 | -0.125 | -0.2672 | No |
| 61 | Npc1 | 8220 | -0.130 | -0.2698 | No |
| 62 | Pcx | 8304 | -0.135 | -0.2722 | No |
| 63 | Lonp1 | 8428 | -0.144 | -0.2776 | No |
| 64 | Ccl25 | 8710 | -0.163 | -0.2952 | No |
| 65 | Jup | 8827 | -0.173 | -0.2990 | No |
| 66 | Gart | 9081 | -0.192 | -0.3134 | No |
| 67 | Slc35b1 | 9128 | -0.196 | -0.3109 | No |
| 68 | Mthfd1 | 9262 | -0.205 | -0.3151 | Yes |
| 69 | Arpp19 | 9335 | -0.211 | -0.3142 | Yes |
| 70 | Il1r1 | 9405 | -0.216 | -0.3129 | Yes |
| 71 | Shmt2 | 9411 | -0.216 | -0.3064 | Yes |
| 72 | Smox | 9475 | -0.221 | -0.3045 | Yes |
| 73 | Irf8 | 9495 | -0.223 | -0.2989 | Yes |
| 74 | Slc6a6 | 9632 | -0.234 | -0.3025 | Yes |
| 75 | Tmem97 | 9726 | -0.241 | -0.3023 | Yes |
| 76 | Gstt2 | 9801 | -0.248 | -0.3004 | Yes |
| 77 | Elovl5 | 9911 | -0.255 | -0.3011 | Yes |
| 78 | Tnfrsf1a | 9932 | -0.257 | -0.2945 | Yes |
| 79 | Pdlim5 | 9976 | -0.261 | -0.2896 | Yes |
| 80 | Bphl | 9988 | -0.262 | -0.2821 | Yes |
| 81 | Papss2 | 10049 | -0.267 | -0.2785 | Yes |
| 82 | Csad | 10057 | -0.268 | -0.2705 | Yes |
| 83 | Id2 | 10328 | -0.291 | -0.2831 | Yes |
| 84 | Casp6 | 10426 | -0.301 | -0.2813 | Yes |
| 85 | Tkfc | 10437 | -0.302 | -0.2725 | Yes |
| 86 | Apoe | 10539 | -0.314 | -0.2706 | Yes |
| 87 | Retsat | 10627 | -0.325 | -0.2673 | Yes |
| 88 | Comt | 10644 | -0.326 | -0.2582 | Yes |
| 89 | Acp1 | 10667 | -0.329 | -0.2494 | Yes |
| 90 | Ptgr1 | 10787 | -0.343 | -0.2481 | Yes |
| 91 | Acox3 | 10799 | -0.344 | -0.2380 | Yes |
| 92 | Cyb5a | 10908 | -0.360 | -0.2353 | Yes |
| 93 | Abcd2 | 10948 | -0.364 | -0.2268 | Yes |
| 94 | Xdh | 10987 | -0.371 | -0.2180 | Yes |
| 95 | Hes6 | 11103 | -0.388 | -0.2150 | Yes |
| 96 | Cndp2 | 11217 | -0.406 | -0.2112 | Yes |
| 97 | Ddt | 11237 | -0.409 | -0.1996 | Yes |
| 98 | Tpst1 | 11268 | -0.413 | -0.1889 | Yes |
| 99 | Nmt1 | 11485 | -0.448 | -0.1921 | Yes |
| 100 | Ssr3 | 11698 | -0.487 | -0.1937 | Yes |
| 101 | Gstm4 | 11732 | -0.494 | -0.1806 | Yes |
| 102 | Cbr1 | 11774 | -0.503 | -0.1678 | Yes |
| 103 | Slc35d1 | 11789 | -0.506 | -0.1528 | Yes |
| 104 | Marchf6 | 11792 | -0.507 | -0.1367 | Yes |
| 105 | Psmb10 | 11941 | -0.546 | -0.1313 | Yes |
| 106 | Ttpa | 12031 | -0.570 | -0.1203 | Yes |
| 107 | Ap4b1 | 12140 | -0.615 | -0.1094 | Yes |
| 108 | Acox1 | 12166 | -0.628 | -0.0914 | Yes |
| 109 | Ets2 | 12189 | -0.644 | -0.0726 | Yes |
| 110 | Gabarapl1 | 12233 | -0.669 | -0.0547 | Yes |
| 111 | Adh5 | 12265 | -0.697 | -0.0349 | Yes |
| 112 | Pros1 | 12288 | -0.716 | -0.0138 | Yes |
| 113 | Dhrs7 | 12370 | -0.818 | 0.0058 | Yes |