Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
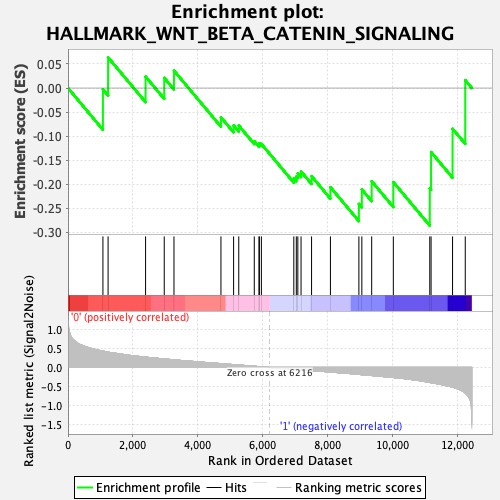
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
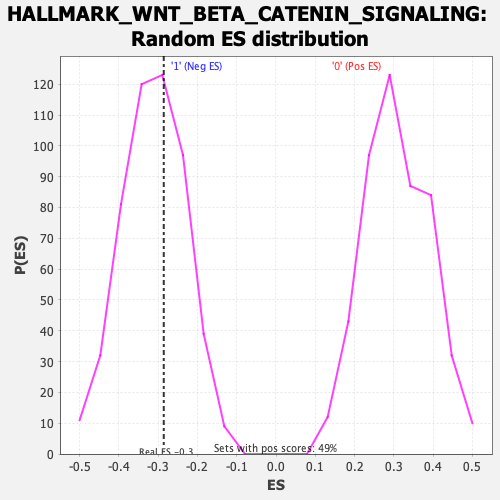
| Enrichment Score (ES) | -0.28572738 |
| Normalized Enrichment Score (NES) | -0.9116429 |
| Nominal p-value | 0.6269531 |
| FDR q-value | 0.8813408 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
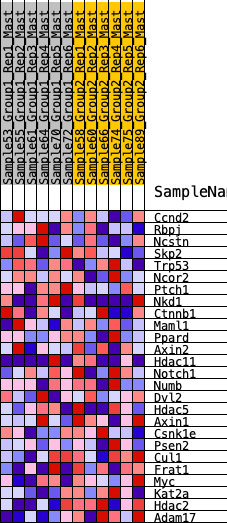
| 1 | Ccnd2 | 1076 | 0.430 | -0.0026 | No |
| 2 | Rbpj | 1235 | 0.404 | 0.0636 | No |
| 3 | Ncstn | 2390 | 0.272 | 0.0238 | No |
| 4 | Skp2 | 2965 | 0.223 | 0.0211 | No |
| 5 | Trp53 | 3265 | 0.200 | 0.0362 | No |
| 6 | Ncor2 | 4712 | 0.100 | -0.0607 | No |
| 7 | Ptch1 | 5102 | 0.073 | -0.0777 | No |
| 8 | Nkd1 | 5261 | 0.064 | -0.0780 | No |
| 9 | Ctnnb1 | 5739 | 0.030 | -0.1104 | No |
| 10 | Maml1 | 5884 | 0.022 | -0.1178 | No |
| 11 | Ppard | 5896 | 0.021 | -0.1146 | No |
| 12 | Axin2 | 5958 | 0.017 | -0.1162 | No |
| 13 | Hdac11 | 6957 | -0.046 | -0.1875 | No |
| 14 | Notch1 | 7034 | -0.050 | -0.1838 | No |
| 15 | Numb | 7081 | -0.053 | -0.1771 | No |
| 16 | Dvl2 | 7180 | -0.060 | -0.1732 | No |
| 17 | Hdac5 | 7504 | -0.083 | -0.1830 | No |
| 18 | Axin1 | 8085 | -0.122 | -0.2059 | No |
| 19 | Csnk1e | 8962 | -0.183 | -0.2407 | Yes |
| 20 | Psen2 | 9053 | -0.190 | -0.2107 | Yes |
| 21 | Cul1 | 9356 | -0.212 | -0.1936 | Yes |
| 22 | Frat1 | 10023 | -0.265 | -0.1954 | Yes |
| 23 | Myc | 11146 | -0.395 | -0.2084 | Yes |
| 24 | Kat2a | 11186 | -0.402 | -0.1330 | Yes |
| 25 | Hdac2 | 11848 | -0.520 | -0.0846 | Yes |
| 26 | Adam17 | 12239 | -0.676 | 0.0163 | Yes |