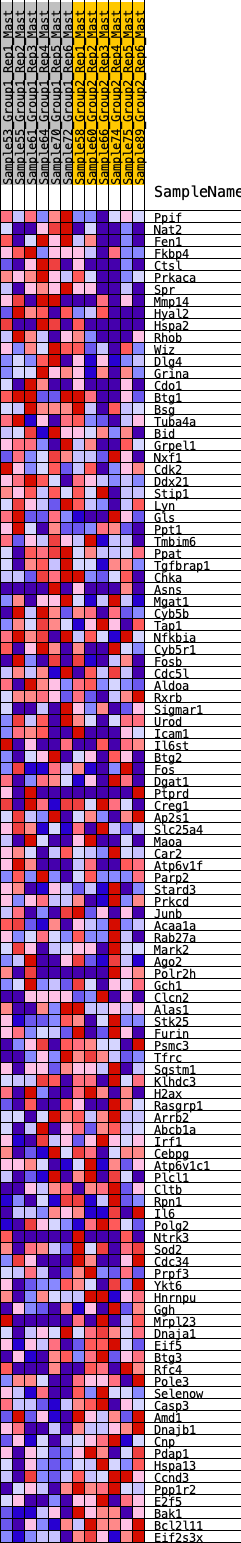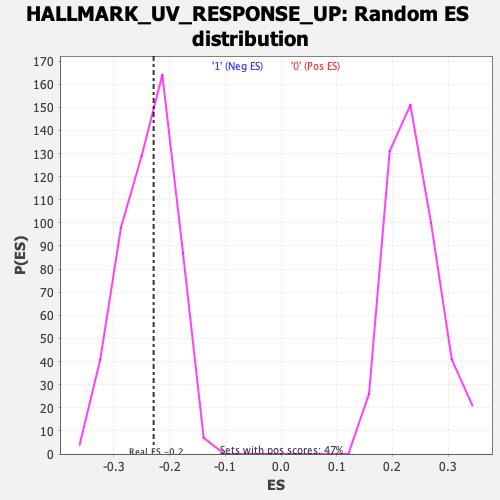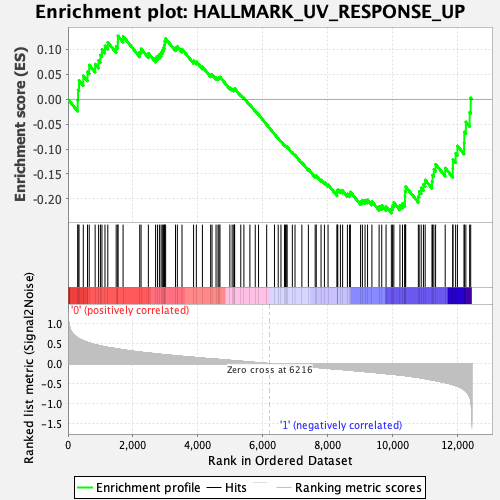
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | -0.22856814 |
| Normalized Enrichment Score (NES) | -0.95913875 |
| Nominal p-value | 0.5377358 |
| FDR q-value | 0.88172835 |
| FWER p-Value | 0.994 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ppif | 300 | 0.635 | -0.0023 | No |
| 2 | Nat2 | 310 | 0.631 | 0.0188 | No |
| 3 | Fen1 | 340 | 0.618 | 0.0379 | No |
| 4 | Fkbp4 | 471 | 0.570 | 0.0471 | No |
| 5 | Ctsl | 601 | 0.533 | 0.0551 | No |
| 6 | Prkaca | 654 | 0.519 | 0.0688 | No |
| 7 | Spr | 836 | 0.476 | 0.0706 | No |
| 8 | Mmp14 | 942 | 0.454 | 0.0778 | No |
| 9 | Hyal2 | 999 | 0.444 | 0.0887 | No |
| 10 | Hspa2 | 1046 | 0.435 | 0.1000 | No |
| 11 | Rhob | 1139 | 0.420 | 0.1071 | No |
| 12 | Wiz | 1226 | 0.405 | 0.1141 | No |
| 13 | Dlg4 | 1484 | 0.371 | 0.1062 | No |
| 14 | Grina | 1536 | 0.363 | 0.1146 | No |
| 15 | Cdo1 | 1538 | 0.363 | 0.1271 | No |
| 16 | Btg1 | 1697 | 0.344 | 0.1262 | No |
| 17 | Bsg | 2210 | 0.289 | 0.0947 | No |
| 18 | Tuba4a | 2251 | 0.284 | 0.1013 | No |
| 19 | Bid | 2476 | 0.264 | 0.0923 | No |
| 20 | Grpel1 | 2698 | 0.246 | 0.0829 | No |
| 21 | Nxf1 | 2756 | 0.241 | 0.0866 | No |
| 22 | Cdk2 | 2816 | 0.235 | 0.0900 | No |
| 23 | Ddx21 | 2868 | 0.231 | 0.0938 | No |
| 24 | Stip1 | 2906 | 0.227 | 0.0987 | No |
| 25 | Lyn | 2944 | 0.224 | 0.1034 | No |
| 26 | Gls | 2973 | 0.222 | 0.1089 | No |
| 27 | Ppt1 | 2979 | 0.222 | 0.1161 | No |
| 28 | Tmbim6 | 3008 | 0.220 | 0.1215 | No |
| 29 | Ppat | 3312 | 0.196 | 0.1037 | No |
| 30 | Tgfbrap1 | 3369 | 0.192 | 0.1058 | No |
| 31 | Chka | 3512 | 0.182 | 0.1006 | No |
| 32 | Asns | 3869 | 0.157 | 0.0772 | No |
| 33 | Mgat1 | 3954 | 0.150 | 0.0756 | No |
| 34 | Cyb5b | 4141 | 0.138 | 0.0653 | No |
| 35 | Tap1 | 4395 | 0.121 | 0.0489 | No |
| 36 | Nfkbia | 4437 | 0.118 | 0.0497 | No |
| 37 | Cyb5r1 | 4561 | 0.110 | 0.0435 | No |
| 38 | Fosb | 4622 | 0.106 | 0.0423 | No |
| 39 | Cdc5l | 4652 | 0.104 | 0.0436 | No |
| 40 | Aldoa | 4682 | 0.102 | 0.0448 | No |
| 41 | Rxrb | 4987 | 0.082 | 0.0230 | No |
| 42 | Sigmar1 | 5057 | 0.076 | 0.0200 | No |
| 43 | Urod | 5107 | 0.073 | 0.0186 | No |
| 44 | Icam1 | 5130 | 0.072 | 0.0193 | No |
| 45 | Il6st | 5134 | 0.072 | 0.0215 | No |
| 46 | Btg2 | 5323 | 0.060 | 0.0084 | No |
| 47 | Fos | 5419 | 0.053 | 0.0025 | No |
| 48 | Dgat1 | 5604 | 0.040 | -0.0110 | No |
| 49 | Ptprd | 5769 | 0.029 | -0.0233 | No |
| 50 | Creg1 | 5870 | 0.022 | -0.0307 | No |
| 51 | Ap2s1 | 6121 | 0.006 | -0.0507 | No |
| 52 | Slc25a4 | 6361 | -0.009 | -0.0698 | No |
| 53 | Maoa | 6476 | -0.016 | -0.0785 | No |
| 54 | Car2 | 6561 | -0.023 | -0.0845 | No |
| 55 | Atp6v1f | 6669 | -0.028 | -0.0922 | No |
| 56 | Parp2 | 6700 | -0.030 | -0.0936 | No |
| 57 | Stard3 | 6725 | -0.031 | -0.0945 | No |
| 58 | Prkcd | 6752 | -0.033 | -0.0955 | No |
| 59 | Junb | 6912 | -0.042 | -0.1069 | No |
| 60 | Acaa1a | 6994 | -0.048 | -0.1118 | No |
| 61 | Rab27a | 7205 | -0.062 | -0.1267 | No |
| 62 | Mark2 | 7406 | -0.076 | -0.1403 | No |
| 63 | Ago2 | 7615 | -0.090 | -0.1540 | No |
| 64 | Polr2h | 7649 | -0.092 | -0.1535 | No |
| 65 | Gch1 | 7796 | -0.100 | -0.1619 | No |
| 66 | Clcn2 | 7900 | -0.108 | -0.1665 | No |
| 67 | Alas1 | 8012 | -0.117 | -0.1715 | No |
| 68 | Stk25 | 8290 | -0.134 | -0.1893 | No |
| 69 | Furin | 8292 | -0.134 | -0.1847 | No |
| 70 | Psmc3 | 8312 | -0.136 | -0.1815 | No |
| 71 | Tfrc | 8393 | -0.142 | -0.1831 | No |
| 72 | Sqstm1 | 8461 | -0.146 | -0.1835 | No |
| 73 | Klhdc3 | 8608 | -0.156 | -0.1899 | No |
| 74 | H2ax | 8674 | -0.160 | -0.1896 | No |
| 75 | Rasgrp1 | 8700 | -0.162 | -0.1860 | No |
| 76 | Arrb2 | 9014 | -0.187 | -0.2049 | No |
| 77 | Abcb1a | 9070 | -0.192 | -0.2028 | No |
| 78 | Irf1 | 9152 | -0.197 | -0.2025 | No |
| 79 | Cebpg | 9229 | -0.203 | -0.2016 | No |
| 80 | Atp6v1c1 | 9363 | -0.213 | -0.2050 | No |
| 81 | Plcl1 | 9585 | -0.231 | -0.2150 | No |
| 82 | Cltb | 9666 | -0.236 | -0.2133 | No |
| 83 | Rpn1 | 9799 | -0.247 | -0.2154 | No |
| 84 | Il6 | 9962 | -0.260 | -0.2196 | Yes |
| 85 | Polg2 | 10000 | -0.263 | -0.2134 | Yes |
| 86 | Ntrk3 | 10039 | -0.267 | -0.2073 | Yes |
| 87 | Sod2 | 10227 | -0.282 | -0.2127 | Yes |
| 88 | Cdc34 | 10306 | -0.289 | -0.2090 | Yes |
| 89 | Prpf3 | 10380 | -0.296 | -0.2046 | Yes |
| 90 | Ykt6 | 10381 | -0.297 | -0.1944 | Yes |
| 91 | Hnrnpu | 10385 | -0.297 | -0.1843 | Yes |
| 92 | Ggh | 10404 | -0.299 | -0.1754 | Yes |
| 93 | Mrpl23 | 10794 | -0.344 | -0.1951 | Yes |
| 94 | Dnaja1 | 10819 | -0.347 | -0.1850 | Yes |
| 95 | Eif5 | 10882 | -0.356 | -0.1777 | Yes |
| 96 | Btg3 | 10951 | -0.365 | -0.1706 | Yes |
| 97 | Rfc4 | 11004 | -0.373 | -0.1619 | Yes |
| 98 | Pole3 | 11214 | -0.405 | -0.1648 | Yes |
| 99 | Selenow | 11232 | -0.409 | -0.1520 | Yes |
| 100 | Casp3 | 11272 | -0.413 | -0.1409 | Yes |
| 101 | Amd1 | 11325 | -0.422 | -0.1305 | Yes |
| 102 | Dnajb1 | 11621 | -0.474 | -0.1380 | Yes |
| 103 | Cnp | 11855 | -0.522 | -0.1388 | Yes |
| 104 | Pdap1 | 11859 | -0.523 | -0.1209 | Yes |
| 105 | Hspa13 | 11942 | -0.546 | -0.1087 | Yes |
| 106 | Ccnd3 | 11998 | -0.562 | -0.0937 | Yes |
| 107 | Ppp1r2 | 12203 | -0.650 | -0.0877 | Yes |
| 108 | E2f5 | 12210 | -0.654 | -0.0655 | Yes |
| 109 | Bak1 | 12258 | -0.690 | -0.0455 | Yes |
| 110 | Bcl2l11 | 12374 | -0.827 | -0.0262 | Yes |
| 111 | Eif2s3x | 12407 | -0.910 | 0.0028 | Yes |