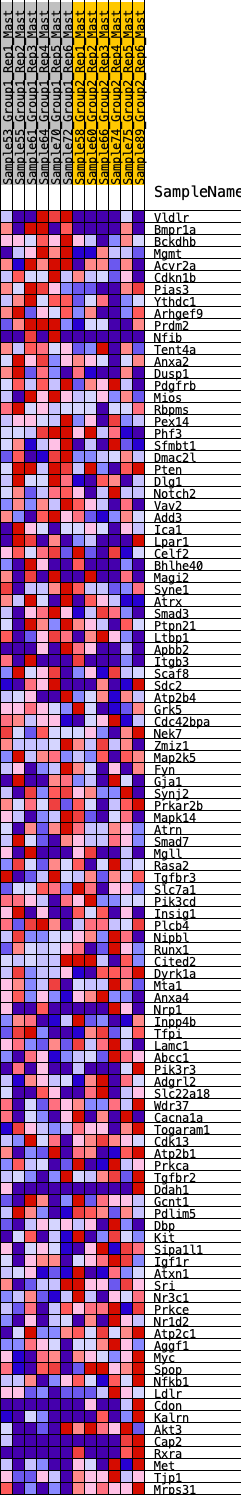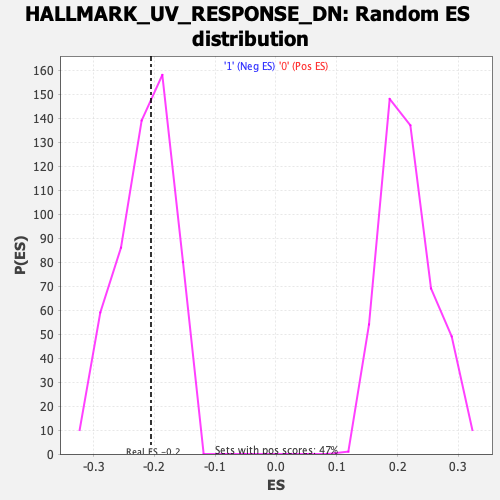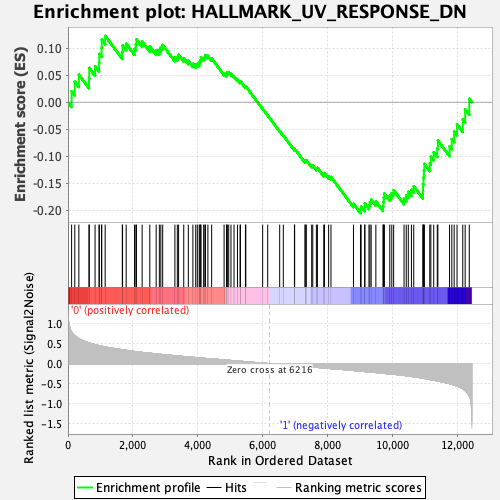
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | -0.20575649 |
| Normalized Enrichment Score (NES) | -0.9526425 |
| Nominal p-value | 0.5413534 |
| FDR q-value | 0.8492747 |
| FWER p-Value | 0.994 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vldlr | 111 | 0.790 | 0.0202 | No |
| 2 | Bmpr1a | 209 | 0.700 | 0.0382 | No |
| 3 | Bckdhb | 335 | 0.620 | 0.0509 | No |
| 4 | Mgmt | 647 | 0.520 | 0.0449 | No |
| 5 | Acvr2a | 655 | 0.518 | 0.0635 | No |
| 6 | Cdkn1b | 834 | 0.476 | 0.0667 | No |
| 7 | Pias3 | 957 | 0.451 | 0.0734 | No |
| 8 | Ythdc1 | 964 | 0.450 | 0.0896 | No |
| 9 | Arhgef9 | 1033 | 0.438 | 0.1002 | No |
| 10 | Prdm2 | 1040 | 0.437 | 0.1159 | No |
| 11 | Nfib | 1145 | 0.419 | 0.1229 | No |
| 12 | Tent4a | 1673 | 0.346 | 0.0930 | No |
| 13 | Anxa2 | 1681 | 0.345 | 0.1052 | No |
| 14 | Dusp1 | 1792 | 0.333 | 0.1085 | No |
| 15 | Pdgfrb | 2050 | 0.305 | 0.0989 | No |
| 16 | Mios | 2091 | 0.300 | 0.1068 | No |
| 17 | Rbpms | 2109 | 0.299 | 0.1164 | No |
| 18 | Pex14 | 2284 | 0.281 | 0.1127 | No |
| 19 | Phf3 | 2520 | 0.260 | 0.1033 | No |
| 20 | Sfmbt1 | 2716 | 0.244 | 0.0965 | No |
| 21 | Dmac2l | 2814 | 0.236 | 0.0973 | No |
| 22 | Pten | 2860 | 0.231 | 0.1022 | No |
| 23 | Dlg1 | 2917 | 0.226 | 0.1060 | No |
| 24 | Notch2 | 3293 | 0.199 | 0.0830 | No |
| 25 | Vav2 | 3368 | 0.192 | 0.0840 | No |
| 26 | Add3 | 3407 | 0.190 | 0.0880 | No |
| 27 | Ica1 | 3566 | 0.177 | 0.0817 | No |
| 28 | Lpar1 | 3707 | 0.169 | 0.0766 | No |
| 29 | Celf2 | 3848 | 0.159 | 0.0711 | No |
| 30 | Bhlhe40 | 3935 | 0.151 | 0.0697 | No |
| 31 | Magi2 | 3984 | 0.148 | 0.0713 | No |
| 32 | Syne1 | 4042 | 0.144 | 0.0720 | No |
| 33 | Atrx | 4062 | 0.143 | 0.0757 | No |
| 34 | Smad3 | 4088 | 0.142 | 0.0790 | No |
| 35 | Ptpn21 | 4096 | 0.142 | 0.0836 | No |
| 36 | Ltbp1 | 4179 | 0.136 | 0.0820 | No |
| 37 | Apbb2 | 4217 | 0.132 | 0.0838 | No |
| 38 | Itgb3 | 4234 | 0.130 | 0.0874 | No |
| 39 | Scaf8 | 4310 | 0.126 | 0.0859 | No |
| 40 | Sdc2 | 4426 | 0.119 | 0.0810 | No |
| 41 | Atp2b4 | 4809 | 0.094 | 0.0535 | No |
| 42 | Grk5 | 4885 | 0.090 | 0.0507 | No |
| 43 | Cdc42bpa | 4890 | 0.090 | 0.0537 | No |
| 44 | Nek7 | 4908 | 0.088 | 0.0556 | No |
| 45 | Zmiz1 | 4951 | 0.084 | 0.0553 | No |
| 46 | Map2k5 | 5021 | 0.080 | 0.0526 | No |
| 47 | Fyn | 5115 | 0.073 | 0.0478 | No |
| 48 | Gja1 | 5221 | 0.066 | 0.0417 | No |
| 49 | Synj2 | 5302 | 0.061 | 0.0375 | No |
| 50 | Prkar2b | 5312 | 0.060 | 0.0390 | No |
| 51 | Mapk14 | 5467 | 0.049 | 0.0283 | No |
| 52 | Atrn | 5478 | 0.048 | 0.0293 | No |
| 53 | Smad7 | 5997 | 0.015 | -0.0122 | No |
| 54 | Mgll | 6153 | 0.004 | -0.0246 | No |
| 55 | Rasa2 | 6522 | -0.019 | -0.0537 | No |
| 56 | Tgfbr3 | 6633 | -0.027 | -0.0617 | No |
| 57 | Slc7a1 | 6976 | -0.047 | -0.0877 | No |
| 58 | Pik3cd | 6983 | -0.047 | -0.0864 | No |
| 59 | Insig1 | 7304 | -0.069 | -0.1098 | No |
| 60 | Plcb4 | 7307 | -0.069 | -0.1074 | No |
| 61 | Nipbl | 7343 | -0.072 | -0.1076 | No |
| 62 | Runx1 | 7508 | -0.083 | -0.1178 | No |
| 63 | Cited2 | 7541 | -0.086 | -0.1173 | No |
| 64 | Dyrk1a | 7652 | -0.092 | -0.1228 | No |
| 65 | Mta1 | 7684 | -0.094 | -0.1218 | No |
| 66 | Anxa4 | 7880 | -0.106 | -0.1337 | No |
| 67 | Nrp1 | 7901 | -0.108 | -0.1313 | No |
| 68 | Inpp4b | 8030 | -0.118 | -0.1373 | No |
| 69 | Tfpi | 8102 | -0.123 | -0.1385 | No |
| 70 | Lamc1 | 8795 | -0.171 | -0.1883 | No |
| 71 | Abcc1 | 9011 | -0.187 | -0.1989 | Yes |
| 72 | Pik3r3 | 9031 | -0.189 | -0.1934 | Yes |
| 73 | Adgrl2 | 9143 | -0.197 | -0.1952 | Yes |
| 74 | Slc22a18 | 9145 | -0.197 | -0.1880 | Yes |
| 75 | Wdr37 | 9273 | -0.206 | -0.1907 | Yes |
| 76 | Cacna1a | 9302 | -0.208 | -0.1852 | Yes |
| 77 | Togaram1 | 9340 | -0.211 | -0.1804 | Yes |
| 78 | Cdk13 | 9485 | -0.222 | -0.1839 | Yes |
| 79 | Atp2b1 | 9705 | -0.239 | -0.1928 | Yes |
| 80 | Prkca | 9717 | -0.241 | -0.1848 | Yes |
| 81 | Tgfbr2 | 9730 | -0.242 | -0.1769 | Yes |
| 82 | Ddah1 | 9747 | -0.243 | -0.1692 | Yes |
| 83 | Gcnt1 | 9914 | -0.255 | -0.1732 | Yes |
| 84 | Pdlim5 | 9976 | -0.261 | -0.1685 | Yes |
| 85 | Dbp | 10033 | -0.266 | -0.1632 | Yes |
| 86 | Kit | 10353 | -0.294 | -0.1782 | Yes |
| 87 | Sipa1l1 | 10421 | -0.301 | -0.1726 | Yes |
| 88 | Igf1r | 10484 | -0.307 | -0.1662 | Yes |
| 89 | Atxn1 | 10580 | -0.319 | -0.1622 | Yes |
| 90 | Sri | 10652 | -0.327 | -0.1558 | Yes |
| 91 | Nr3c1 | 10936 | -0.363 | -0.1653 | Yes |
| 92 | Prkce | 10939 | -0.363 | -0.1521 | Yes |
| 93 | Nr1d2 | 10953 | -0.365 | -0.1397 | Yes |
| 94 | Atp2c1 | 10965 | -0.367 | -0.1270 | Yes |
| 95 | Aggf1 | 10979 | -0.369 | -0.1144 | Yes |
| 96 | Myc | 11146 | -0.395 | -0.1133 | Yes |
| 97 | Spop | 11179 | -0.401 | -0.1011 | Yes |
| 98 | Nfkb1 | 11267 | -0.413 | -0.0929 | Yes |
| 99 | Ldlr | 11377 | -0.431 | -0.0858 | Yes |
| 100 | Cdon | 11396 | -0.434 | -0.0712 | Yes |
| 101 | Kalrn | 11755 | -0.498 | -0.0819 | Yes |
| 102 | Akt3 | 11825 | -0.515 | -0.0684 | Yes |
| 103 | Cap2 | 11897 | -0.532 | -0.0546 | Yes |
| 104 | Rxra | 11986 | -0.559 | -0.0411 | Yes |
| 105 | Met | 12162 | -0.624 | -0.0322 | Yes |
| 106 | Tjp1 | 12236 | -0.672 | -0.0133 | Yes |
| 107 | Mrps31 | 12363 | -0.808 | 0.0063 | Yes |