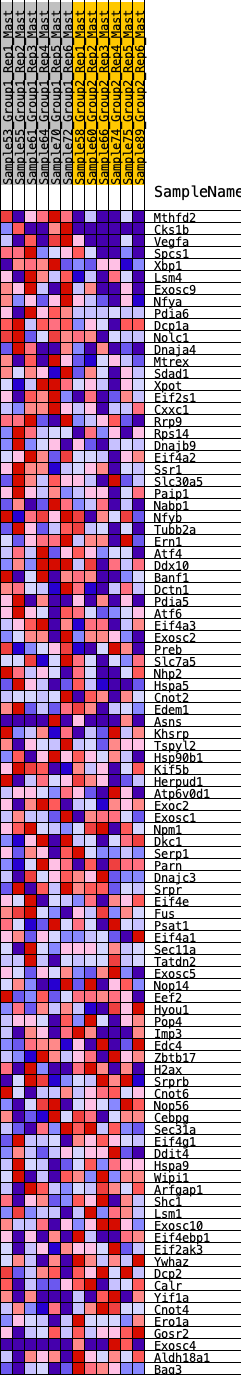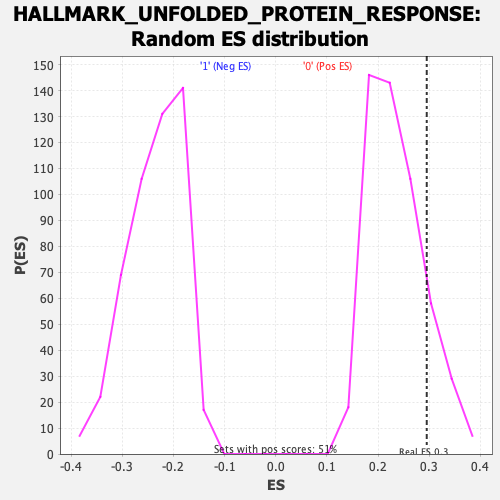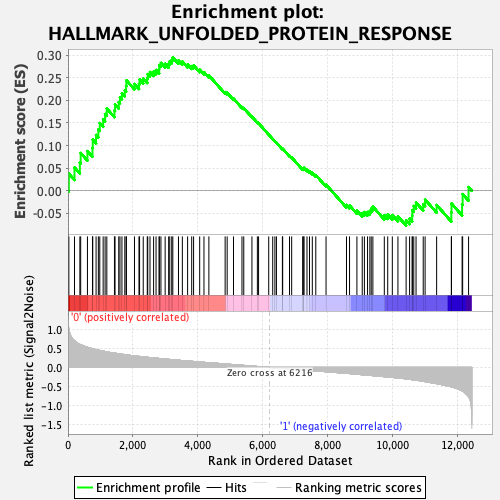
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.29516682 |
| Normalized Enrichment Score (NES) | 1.2609621 |
| Nominal p-value | 0.14003944 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.804 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mthfd2 | 26 | 1.015 | 0.0377 | Yes |
| 2 | Cks1b | 200 | 0.707 | 0.0515 | Yes |
| 3 | Vegfa | 366 | 0.607 | 0.0619 | Yes |
| 4 | Spcs1 | 391 | 0.600 | 0.0836 | Yes |
| 5 | Xbp1 | 599 | 0.535 | 0.0878 | Yes |
| 6 | Lsm4 | 754 | 0.492 | 0.0946 | Yes |
| 7 | Exosc9 | 763 | 0.490 | 0.1132 | Yes |
| 8 | Nfya | 865 | 0.470 | 0.1235 | Yes |
| 9 | Pdia6 | 935 | 0.455 | 0.1357 | Yes |
| 10 | Dcp1a | 979 | 0.448 | 0.1498 | Yes |
| 11 | Nolc1 | 1083 | 0.428 | 0.1583 | Yes |
| 12 | Dnaja4 | 1143 | 0.419 | 0.1700 | Yes |
| 13 | Mtrex | 1196 | 0.410 | 0.1819 | Yes |
| 14 | Sdad1 | 1431 | 0.378 | 0.1778 | Yes |
| 15 | Xpot | 1449 | 0.377 | 0.1912 | Yes |
| 16 | Eif2s1 | 1563 | 0.361 | 0.1962 | Yes |
| 17 | Cxxc1 | 1601 | 0.357 | 0.2072 | Yes |
| 18 | Rrp9 | 1662 | 0.347 | 0.2159 | Yes |
| 19 | Rps14 | 1750 | 0.338 | 0.2222 | Yes |
| 20 | Dnajb9 | 1791 | 0.333 | 0.2320 | Yes |
| 21 | Eif4a2 | 1800 | 0.332 | 0.2444 | Yes |
| 22 | Ssr1 | 2048 | 0.305 | 0.2363 | Yes |
| 23 | Slc30a5 | 2187 | 0.291 | 0.2365 | Yes |
| 24 | Paip1 | 2205 | 0.289 | 0.2465 | Yes |
| 25 | Nabp1 | 2318 | 0.278 | 0.2484 | Yes |
| 26 | Nfyb | 2442 | 0.267 | 0.2489 | Yes |
| 27 | Tubb2a | 2457 | 0.266 | 0.2582 | Yes |
| 28 | Ern1 | 2526 | 0.260 | 0.2629 | Yes |
| 29 | Atf4 | 2637 | 0.251 | 0.2638 | Yes |
| 30 | Ddx10 | 2713 | 0.244 | 0.2673 | Yes |
| 31 | Banf1 | 2809 | 0.236 | 0.2689 | Yes |
| 32 | Dctn1 | 2811 | 0.236 | 0.2781 | Yes |
| 33 | Pdia5 | 2863 | 0.231 | 0.2830 | Yes |
| 34 | Atf6 | 2993 | 0.221 | 0.2813 | Yes |
| 35 | Eif4a3 | 3101 | 0.213 | 0.2810 | Yes |
| 36 | Exosc2 | 3135 | 0.210 | 0.2865 | Yes |
| 37 | Preb | 3196 | 0.206 | 0.2898 | Yes |
| 38 | Slc7a5 | 3229 | 0.203 | 0.2952 | Yes |
| 39 | Nhp2 | 3404 | 0.190 | 0.2885 | No |
| 40 | Hspa5 | 3523 | 0.181 | 0.2861 | No |
| 41 | Cnot2 | 3685 | 0.170 | 0.2797 | No |
| 42 | Edem1 | 3811 | 0.161 | 0.2759 | No |
| 43 | Asns | 3869 | 0.157 | 0.2774 | No |
| 44 | Khsrp | 4057 | 0.143 | 0.2679 | No |
| 45 | Tspyl2 | 4189 | 0.135 | 0.2626 | No |
| 46 | Hsp90b1 | 4340 | 0.124 | 0.2553 | No |
| 47 | Kif5b | 4841 | 0.092 | 0.2184 | No |
| 48 | Herpud1 | 4904 | 0.088 | 0.2168 | No |
| 49 | Atp6v0d1 | 5099 | 0.073 | 0.2040 | No |
| 50 | Exoc2 | 5360 | 0.057 | 0.1852 | No |
| 51 | Exosc1 | 5415 | 0.053 | 0.1829 | No |
| 52 | Npm1 | 5667 | 0.035 | 0.1640 | No |
| 53 | Dkc1 | 5834 | 0.025 | 0.1515 | No |
| 54 | Serp1 | 5871 | 0.022 | 0.1495 | No |
| 55 | Parn | 6184 | 0.002 | 0.1243 | No |
| 56 | Dnajc3 | 6307 | -0.006 | 0.1146 | No |
| 57 | Srpr | 6374 | -0.010 | 0.1097 | No |
| 58 | Eif4e | 6422 | -0.012 | 0.1064 | No |
| 59 | Fus | 6605 | -0.025 | 0.0926 | No |
| 60 | Psat1 | 6614 | -0.025 | 0.0929 | No |
| 61 | Eif4a1 | 6824 | -0.036 | 0.0774 | No |
| 62 | Sec11a | 6891 | -0.040 | 0.0737 | No |
| 63 | Tatdn2 | 7229 | -0.063 | 0.0488 | No |
| 64 | Exosc5 | 7263 | -0.067 | 0.0488 | No |
| 65 | Nop14 | 7269 | -0.067 | 0.0510 | No |
| 66 | Eef2 | 7363 | -0.073 | 0.0463 | No |
| 67 | Hyou1 | 7440 | -0.079 | 0.0433 | No |
| 68 | Pop4 | 7527 | -0.085 | 0.0396 | No |
| 69 | Imp3 | 7634 | -0.092 | 0.0346 | No |
| 70 | Edc4 | 7950 | -0.112 | 0.0135 | No |
| 71 | Zbtb17 | 8580 | -0.154 | -0.0314 | No |
| 72 | H2ax | 8674 | -0.160 | -0.0326 | No |
| 73 | Srprb | 8901 | -0.178 | -0.0439 | No |
| 74 | Cnot6 | 9065 | -0.191 | -0.0496 | No |
| 75 | Nop56 | 9131 | -0.196 | -0.0472 | No |
| 76 | Cebpg | 9229 | -0.203 | -0.0470 | No |
| 77 | Sec31a | 9300 | -0.208 | -0.0446 | No |
| 78 | Eif4g1 | 9342 | -0.211 | -0.0396 | No |
| 79 | Ddit4 | 9393 | -0.215 | -0.0352 | No |
| 80 | Hspa9 | 9745 | -0.243 | -0.0541 | No |
| 81 | Wipi1 | 9850 | -0.251 | -0.0527 | No |
| 82 | Arfgap1 | 9992 | -0.263 | -0.0538 | No |
| 83 | Shc1 | 10165 | -0.277 | -0.0568 | No |
| 84 | Lsm1 | 10417 | -0.300 | -0.0654 | No |
| 85 | Exosc10 | 10524 | -0.313 | -0.0617 | No |
| 86 | Eif4ebp1 | 10604 | -0.323 | -0.0554 | No |
| 87 | Eif2ak3 | 10608 | -0.323 | -0.0430 | No |
| 88 | Ywhaz | 10653 | -0.327 | -0.0337 | No |
| 89 | Dcp2 | 10723 | -0.334 | -0.0261 | No |
| 90 | Calr | 10944 | -0.364 | -0.0297 | No |
| 91 | Yif1a | 11002 | -0.373 | -0.0197 | No |
| 92 | Cnot4 | 11357 | -0.428 | -0.0316 | No |
| 93 | Ero1a | 11810 | -0.512 | -0.0481 | No |
| 94 | Gosr2 | 11815 | -0.513 | -0.0283 | No |
| 95 | Exosc4 | 12142 | -0.616 | -0.0305 | No |
| 96 | Aldh18a1 | 12159 | -0.623 | -0.0074 | No |
| 97 | Bag3 | 12341 | -0.767 | 0.0081 | No |