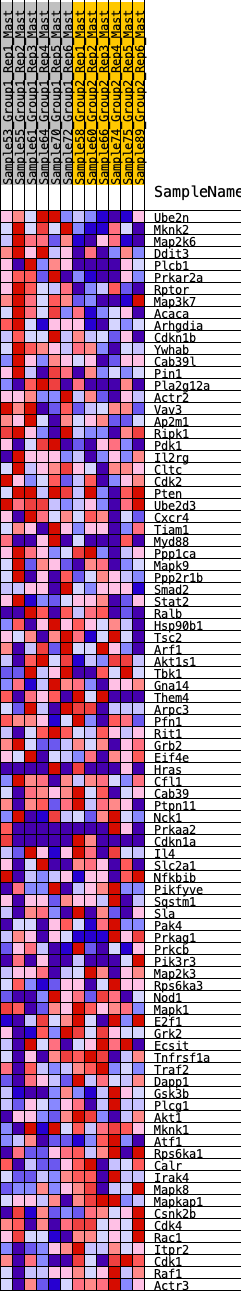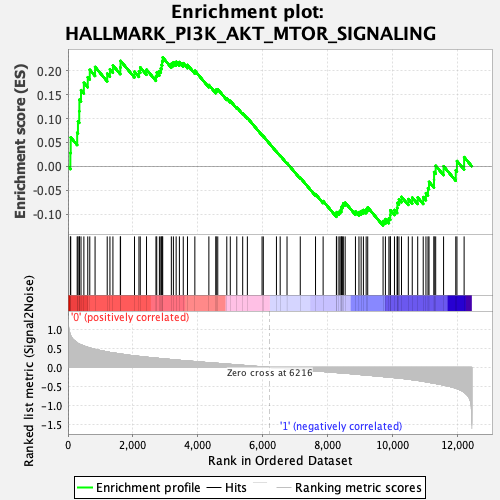
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.22796708 |
| Normalized Enrichment Score (NES) | 1.0150034 |
| Nominal p-value | 0.4642127 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.992 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ube2n | 73 | 0.849 | 0.0280 | Yes |
| 2 | Mknk2 | 84 | 0.830 | 0.0604 | Yes |
| 3 | Map2k6 | 283 | 0.651 | 0.0704 | Yes |
| 4 | Ddit3 | 307 | 0.632 | 0.0939 | Yes |
| 5 | Plcb1 | 346 | 0.614 | 0.1154 | Yes |
| 6 | Prkar2a | 351 | 0.612 | 0.1395 | Yes |
| 7 | Rptor | 401 | 0.597 | 0.1594 | Yes |
| 8 | Map3k7 | 489 | 0.563 | 0.1749 | Yes |
| 9 | Acaca | 609 | 0.529 | 0.1865 | Yes |
| 10 | Arhgdia | 671 | 0.515 | 0.2021 | Yes |
| 11 | Cdkn1b | 834 | 0.476 | 0.2080 | Yes |
| 12 | Ywhab | 1208 | 0.408 | 0.1942 | Yes |
| 13 | Cab39l | 1292 | 0.395 | 0.2033 | Yes |
| 14 | Pin1 | 1384 | 0.384 | 0.2112 | Yes |
| 15 | Pla2g12a | 1611 | 0.355 | 0.2071 | Yes |
| 16 | Actr2 | 1619 | 0.354 | 0.2207 | Yes |
| 17 | Vav3 | 2049 | 0.305 | 0.1982 | Yes |
| 18 | Ap2m1 | 2186 | 0.291 | 0.1988 | Yes |
| 19 | Ripk1 | 2226 | 0.287 | 0.2071 | Yes |
| 20 | Pdk1 | 2419 | 0.269 | 0.2023 | Yes |
| 21 | Il2rg | 2710 | 0.245 | 0.1887 | Yes |
| 22 | Cltc | 2734 | 0.243 | 0.1965 | Yes |
| 23 | Cdk2 | 2816 | 0.235 | 0.1994 | Yes |
| 24 | Pten | 2860 | 0.231 | 0.2051 | Yes |
| 25 | Ube2d3 | 2885 | 0.229 | 0.2123 | Yes |
| 26 | Cxcr4 | 2902 | 0.227 | 0.2201 | Yes |
| 27 | Tiam1 | 2918 | 0.226 | 0.2280 | Yes |
| 28 | Myd88 | 3184 | 0.207 | 0.2148 | No |
| 29 | Ppp1ca | 3247 | 0.202 | 0.2178 | No |
| 30 | Mapk9 | 3332 | 0.195 | 0.2188 | No |
| 31 | Ppp2r1b | 3432 | 0.188 | 0.2183 | No |
| 32 | Smad2 | 3551 | 0.178 | 0.2159 | No |
| 33 | Stat2 | 3681 | 0.170 | 0.2123 | No |
| 34 | Ralb | 3908 | 0.154 | 0.2001 | No |
| 35 | Hsp90b1 | 4340 | 0.124 | 0.1702 | No |
| 36 | Tsc2 | 4555 | 0.110 | 0.1573 | No |
| 37 | Arf1 | 4566 | 0.109 | 0.1608 | No |
| 38 | Akt1s1 | 4614 | 0.107 | 0.1613 | No |
| 39 | Tbk1 | 4892 | 0.089 | 0.1424 | No |
| 40 | Gna14 | 5000 | 0.081 | 0.1370 | No |
| 41 | Them4 | 5200 | 0.067 | 0.1236 | No |
| 42 | Arpc3 | 5382 | 0.056 | 0.1112 | No |
| 43 | Pfn1 | 5526 | 0.045 | 0.1014 | No |
| 44 | Rit1 | 5979 | 0.016 | 0.0655 | No |
| 45 | Grb2 | 6017 | 0.013 | 0.0630 | No |
| 46 | Eif4e | 6422 | -0.012 | 0.0308 | No |
| 47 | Hras | 6537 | -0.021 | 0.0224 | No |
| 48 | Cfl1 | 6748 | -0.032 | 0.0067 | No |
| 49 | Cab39 | 7156 | -0.058 | -0.0240 | No |
| 50 | Ptpn11 | 7626 | -0.091 | -0.0583 | No |
| 51 | Nck1 | 7860 | -0.105 | -0.0729 | No |
| 52 | Prkaa2 | 8269 | -0.133 | -0.1006 | No |
| 53 | Cdkn1a | 8279 | -0.134 | -0.0960 | No |
| 54 | Il4 | 8347 | -0.138 | -0.0959 | No |
| 55 | Slc2a1 | 8382 | -0.141 | -0.0930 | No |
| 56 | Nfkbib | 8419 | -0.143 | -0.0902 | No |
| 57 | Pikfyve | 8429 | -0.144 | -0.0851 | No |
| 58 | Sqstm1 | 8461 | -0.146 | -0.0818 | No |
| 59 | Sla | 8482 | -0.147 | -0.0775 | No |
| 60 | Pak4 | 8539 | -0.151 | -0.0760 | No |
| 61 | Prkag1 | 8856 | -0.175 | -0.0946 | No |
| 62 | Prkcb | 8964 | -0.183 | -0.0959 | No |
| 63 | Pik3r3 | 9031 | -0.189 | -0.0937 | No |
| 64 | Map2k3 | 9101 | -0.194 | -0.0916 | No |
| 65 | Rps6ka3 | 9186 | -0.199 | -0.0904 | No |
| 66 | Nod1 | 9234 | -0.203 | -0.0860 | No |
| 67 | Mapk1 | 9707 | -0.240 | -0.1147 | No |
| 68 | E2f1 | 9779 | -0.245 | -0.1106 | No |
| 69 | Grk2 | 9887 | -0.253 | -0.1091 | No |
| 70 | Ecsit | 9928 | -0.257 | -0.1021 | No |
| 71 | Tnfrsf1a | 9932 | -0.257 | -0.0921 | No |
| 72 | Traf2 | 10058 | -0.268 | -0.0915 | No |
| 73 | Dapp1 | 10140 | -0.275 | -0.0870 | No |
| 74 | Gsk3b | 10149 | -0.276 | -0.0767 | No |
| 75 | Plcg1 | 10196 | -0.279 | -0.0692 | No |
| 76 | Akt1 | 10275 | -0.287 | -0.0641 | No |
| 77 | Mknk1 | 10485 | -0.307 | -0.0687 | No |
| 78 | Atf1 | 10606 | -0.323 | -0.0655 | No |
| 79 | Rps6ka1 | 10774 | -0.341 | -0.0654 | No |
| 80 | Calr | 10944 | -0.364 | -0.0645 | No |
| 81 | Irak4 | 11028 | -0.377 | -0.0562 | No |
| 82 | Mapk8 | 11091 | -0.386 | -0.0458 | No |
| 83 | Mapkap1 | 11122 | -0.390 | -0.0326 | No |
| 84 | Csnk2b | 11276 | -0.414 | -0.0284 | No |
| 85 | Cdk4 | 11279 | -0.414 | -0.0120 | No |
| 86 | Rac1 | 11328 | -0.423 | 0.0010 | No |
| 87 | Itpr2 | 11571 | -0.464 | 0.0000 | No |
| 88 | Cdk1 | 11947 | -0.547 | -0.0085 | No |
| 89 | Raf1 | 11983 | -0.558 | 0.0110 | No |
| 90 | Actr3 | 12204 | -0.650 | 0.0192 | No |