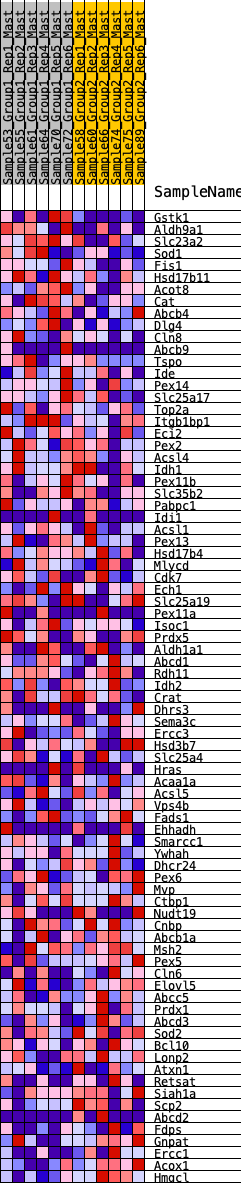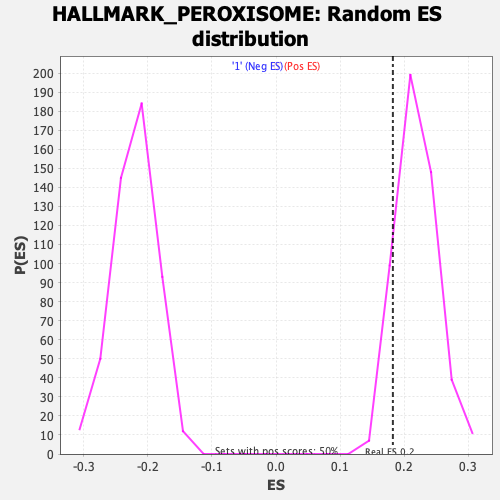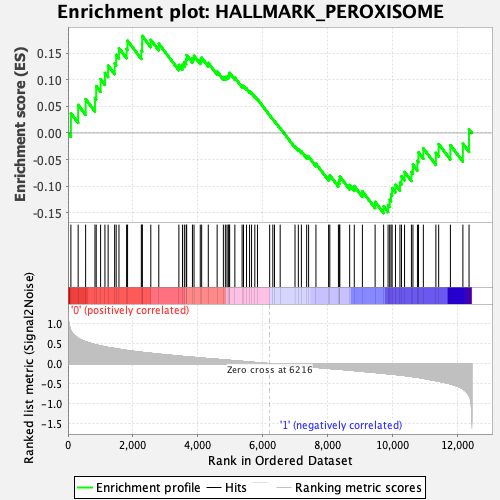
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.18242076 |
| Normalized Enrichment Score (NES) | 0.8325296 |
| Nominal p-value | 0.90656066 |
| FDR q-value | 0.8899058 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gstk1 | 89 | 0.824 | 0.0370 | Yes |
| 2 | Aldh9a1 | 313 | 0.630 | 0.0527 | Yes |
| 3 | Slc23a2 | 543 | 0.548 | 0.0636 | Yes |
| 4 | Sod1 | 831 | 0.477 | 0.0659 | Yes |
| 5 | Fis1 | 869 | 0.469 | 0.0881 | Yes |
| 6 | Hsd17b11 | 1004 | 0.443 | 0.1010 | Yes |
| 7 | Acot8 | 1138 | 0.420 | 0.1128 | Yes |
| 8 | Cat | 1236 | 0.404 | 0.1266 | Yes |
| 9 | Abcb4 | 1437 | 0.378 | 0.1307 | Yes |
| 10 | Dlg4 | 1484 | 0.371 | 0.1469 | Yes |
| 11 | Cln8 | 1570 | 0.360 | 0.1593 | Yes |
| 12 | Abcb9 | 1805 | 0.330 | 0.1581 | Yes |
| 13 | Tspo | 1835 | 0.327 | 0.1733 | Yes |
| 14 | Ide | 2259 | 0.283 | 0.1543 | Yes |
| 15 | Pex14 | 2284 | 0.281 | 0.1674 | Yes |
| 16 | Slc25a17 | 2286 | 0.281 | 0.1824 | Yes |
| 17 | Top2a | 2551 | 0.257 | 0.1749 | No |
| 18 | Itgb1bp1 | 2797 | 0.237 | 0.1678 | No |
| 19 | Eci2 | 3416 | 0.189 | 0.1279 | No |
| 20 | Pex2 | 3527 | 0.180 | 0.1287 | No |
| 21 | Acsl4 | 3585 | 0.176 | 0.1335 | No |
| 22 | Idh1 | 3640 | 0.172 | 0.1384 | No |
| 23 | Pex11b | 3656 | 0.172 | 0.1463 | No |
| 24 | Slc35b2 | 3834 | 0.159 | 0.1406 | No |
| 25 | Pabpc1 | 3883 | 0.156 | 0.1451 | No |
| 26 | Idi1 | 4075 | 0.143 | 0.1373 | No |
| 27 | Acsl1 | 4118 | 0.140 | 0.1414 | No |
| 28 | Pex13 | 4322 | 0.125 | 0.1316 | No |
| 29 | Hsd17b4 | 4595 | 0.107 | 0.1154 | No |
| 30 | Mlycd | 4787 | 0.095 | 0.1050 | No |
| 31 | Cdk7 | 4848 | 0.092 | 0.1051 | No |
| 32 | Ech1 | 4899 | 0.088 | 0.1058 | No |
| 33 | Slc25a19 | 4939 | 0.085 | 0.1072 | No |
| 34 | Pex11a | 4964 | 0.083 | 0.1097 | No |
| 35 | Isoc1 | 4980 | 0.082 | 0.1129 | No |
| 36 | Prdx5 | 5141 | 0.072 | 0.1038 | No |
| 37 | Aldh1a1 | 5364 | 0.056 | 0.0889 | No |
| 38 | Abcd1 | 5407 | 0.054 | 0.0884 | No |
| 39 | Rdh11 | 5500 | 0.047 | 0.0834 | No |
| 40 | Idh2 | 5594 | 0.041 | 0.0781 | No |
| 41 | Crat | 5655 | 0.036 | 0.0752 | No |
| 42 | Dhrs3 | 5756 | 0.030 | 0.0687 | No |
| 43 | Sema3c | 5837 | 0.025 | 0.0636 | No |
| 44 | Ercc3 | 6213 | 0.000 | 0.0332 | No |
| 45 | Hsd3b7 | 6310 | -0.007 | 0.0258 | No |
| 46 | Slc25a4 | 6361 | -0.009 | 0.0223 | No |
| 47 | Hras | 6537 | -0.021 | 0.0092 | No |
| 48 | Acaa1a | 6994 | -0.048 | -0.0251 | No |
| 49 | Acsl5 | 7097 | -0.055 | -0.0304 | No |
| 50 | Vps4b | 7190 | -0.061 | -0.0346 | No |
| 51 | Fads1 | 7353 | -0.072 | -0.0438 | No |
| 52 | Ehhadh | 7413 | -0.077 | -0.0445 | No |
| 53 | Smarcc1 | 7639 | -0.092 | -0.0578 | No |
| 54 | Ywhah | 8029 | -0.118 | -0.0829 | No |
| 55 | Dhcr24 | 8066 | -0.120 | -0.0794 | No |
| 56 | Pex6 | 8331 | -0.137 | -0.0934 | No |
| 57 | Mvp | 8360 | -0.139 | -0.0882 | No |
| 58 | Ctbp1 | 8375 | -0.141 | -0.0817 | No |
| 59 | Nudt19 | 8678 | -0.160 | -0.0976 | No |
| 60 | Cnbp | 8820 | -0.173 | -0.0997 | No |
| 61 | Abcb1a | 9070 | -0.192 | -0.1096 | No |
| 62 | Msh2 | 9462 | -0.220 | -0.1294 | No |
| 63 | Pex5 | 9724 | -0.241 | -0.1376 | No |
| 64 | Cln6 | 9861 | -0.252 | -0.1351 | No |
| 65 | Elovl5 | 9911 | -0.255 | -0.1254 | No |
| 66 | Abcc5 | 9952 | -0.259 | -0.1147 | No |
| 67 | Prdx1 | 9986 | -0.262 | -0.1034 | No |
| 68 | Abcd3 | 10089 | -0.270 | -0.0971 | No |
| 69 | Sod2 | 10227 | -0.282 | -0.0931 | No |
| 70 | Bcl10 | 10274 | -0.287 | -0.0814 | No |
| 71 | Lonp2 | 10369 | -0.295 | -0.0732 | No |
| 72 | Atxn1 | 10580 | -0.319 | -0.0731 | No |
| 73 | Retsat | 10627 | -0.325 | -0.0594 | No |
| 74 | Siah1a | 10766 | -0.340 | -0.0523 | No |
| 75 | Scp2 | 10798 | -0.344 | -0.0364 | No |
| 76 | Abcd2 | 10948 | -0.364 | -0.0289 | No |
| 77 | Fdps | 11334 | -0.423 | -0.0373 | No |
| 78 | Gnpat | 11420 | -0.437 | -0.0208 | No |
| 79 | Ercc1 | 11781 | -0.504 | -0.0228 | No |
| 80 | Acox1 | 12166 | -0.628 | -0.0202 | No |
| 81 | Hmgcl | 12356 | -0.790 | 0.0069 | No |