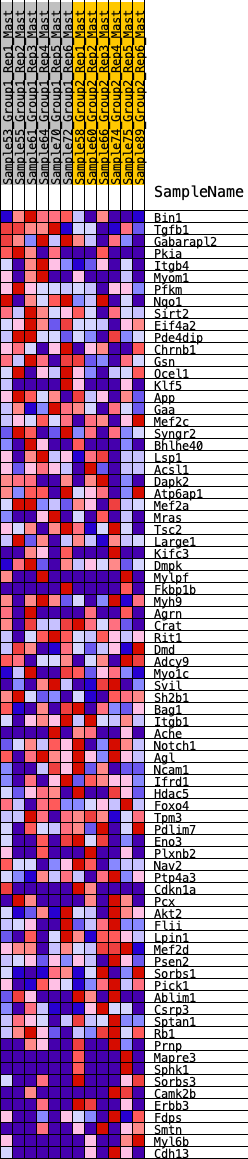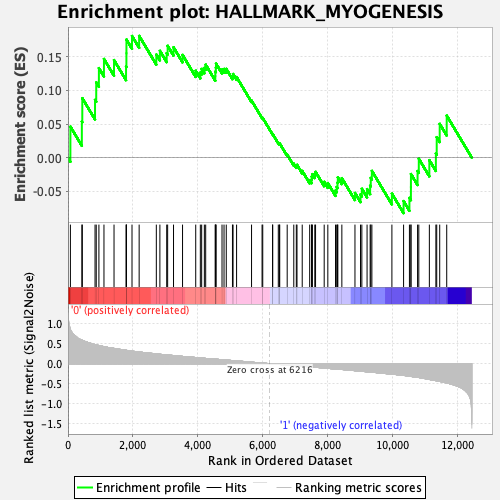
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.18093972 |
| Normalized Enrichment Score (NES) | 0.71298504 |
| Nominal p-value | 0.91803277 |
| FDR q-value | 0.93067145 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bin1 | 74 | 0.849 | 0.0461 | Yes |
| 2 | Tgfb1 | 427 | 0.586 | 0.0537 | Yes |
| 3 | Gabarapl2 | 441 | 0.582 | 0.0884 | Yes |
| 4 | Pkia | 833 | 0.476 | 0.0860 | Yes |
| 5 | Itgb4 | 870 | 0.468 | 0.1119 | Yes |
| 6 | Myom1 | 952 | 0.452 | 0.1331 | Yes |
| 7 | Pfkm | 1108 | 0.425 | 0.1467 | Yes |
| 8 | Nqo1 | 1418 | 0.380 | 0.1450 | Yes |
| 9 | Sirt2 | 1789 | 0.333 | 0.1355 | Yes |
| 10 | Eif4a2 | 1800 | 0.332 | 0.1551 | Yes |
| 11 | Pde4dip | 1801 | 0.331 | 0.1755 | Yes |
| 12 | Chrnb1 | 1972 | 0.311 | 0.1808 | Yes |
| 13 | Gsn | 2192 | 0.290 | 0.1809 | Yes |
| 14 | Ocel1 | 2721 | 0.244 | 0.1532 | No |
| 15 | Klf5 | 2831 | 0.234 | 0.1588 | No |
| 16 | App | 3041 | 0.217 | 0.1552 | No |
| 17 | Gaa | 3070 | 0.216 | 0.1662 | No |
| 18 | Mef2c | 3251 | 0.201 | 0.1640 | No |
| 19 | Syngr2 | 3529 | 0.180 | 0.1527 | No |
| 20 | Bhlhe40 | 3935 | 0.151 | 0.1292 | No |
| 21 | Lsp1 | 4077 | 0.143 | 0.1266 | No |
| 22 | Acsl1 | 4118 | 0.140 | 0.1319 | No |
| 23 | Dapk2 | 4201 | 0.133 | 0.1335 | No |
| 24 | Atp6ap1 | 4242 | 0.129 | 0.1382 | No |
| 25 | Mef2a | 4538 | 0.111 | 0.1211 | No |
| 26 | Mras | 4539 | 0.111 | 0.1279 | No |
| 27 | Tsc2 | 4555 | 0.110 | 0.1335 | No |
| 28 | Large1 | 4560 | 0.110 | 0.1399 | No |
| 29 | Kifc3 | 4745 | 0.098 | 0.1310 | No |
| 30 | Dmpk | 4807 | 0.094 | 0.1319 | No |
| 31 | Mylpf | 4876 | 0.090 | 0.1319 | No |
| 32 | Fkbp1b | 5067 | 0.075 | 0.1212 | No |
| 33 | Myh9 | 5085 | 0.074 | 0.1244 | No |
| 34 | Agrn | 5194 | 0.068 | 0.1198 | No |
| 35 | Crat | 5655 | 0.036 | 0.0848 | No |
| 36 | Rit1 | 5979 | 0.016 | 0.0597 | No |
| 37 | Dmd | 6000 | 0.014 | 0.0589 | No |
| 38 | Adcy9 | 6304 | -0.006 | 0.0348 | No |
| 39 | Myo1c | 6305 | -0.006 | 0.0352 | No |
| 40 | Svil | 6479 | -0.016 | 0.0221 | No |
| 41 | Sh2b1 | 6513 | -0.019 | 0.0206 | No |
| 42 | Bag1 | 6523 | -0.019 | 0.0211 | No |
| 43 | Itgb1 | 6753 | -0.033 | 0.0046 | No |
| 44 | Ache | 6956 | -0.046 | -0.0089 | No |
| 45 | Notch1 | 7034 | -0.050 | -0.0121 | No |
| 46 | Agl | 7055 | -0.052 | -0.0105 | No |
| 47 | Ncam1 | 7216 | -0.063 | -0.0196 | No |
| 48 | Ifrd1 | 7446 | -0.079 | -0.0333 | No |
| 49 | Hdac5 | 7504 | -0.083 | -0.0328 | No |
| 50 | Foxo4 | 7509 | -0.083 | -0.0280 | No |
| 51 | Tpm3 | 7530 | -0.085 | -0.0244 | No |
| 52 | Pdlim7 | 7604 | -0.090 | -0.0248 | No |
| 53 | Eno3 | 7627 | -0.091 | -0.0210 | No |
| 54 | Plxnb2 | 7893 | -0.107 | -0.0358 | No |
| 55 | Nav2 | 8005 | -0.116 | -0.0377 | No |
| 56 | Ptp4a3 | 8242 | -0.132 | -0.0487 | No |
| 57 | Cdkn1a | 8279 | -0.134 | -0.0434 | No |
| 58 | Pcx | 8304 | -0.135 | -0.0370 | No |
| 59 | Akt2 | 8313 | -0.136 | -0.0293 | No |
| 60 | Flii | 8437 | -0.145 | -0.0304 | No |
| 61 | Lpin1 | 8841 | -0.174 | -0.0523 | No |
| 62 | Mef2d | 9012 | -0.187 | -0.0546 | No |
| 63 | Psen2 | 9053 | -0.190 | -0.0461 | No |
| 64 | Sorbs1 | 9214 | -0.202 | -0.0466 | No |
| 65 | Pick1 | 9309 | -0.209 | -0.0414 | No |
| 66 | Ablim1 | 9328 | -0.210 | -0.0299 | No |
| 67 | Csrp3 | 9362 | -0.213 | -0.0195 | No |
| 68 | Sptan1 | 9979 | -0.261 | -0.0533 | No |
| 69 | Rb1 | 10339 | -0.293 | -0.0644 | No |
| 70 | Prnp | 10519 | -0.312 | -0.0597 | No |
| 71 | Mapre3 | 10565 | -0.318 | -0.0438 | No |
| 72 | Sphk1 | 10566 | -0.318 | -0.0242 | No |
| 73 | Sorbs3 | 10770 | -0.340 | -0.0197 | No |
| 74 | Camk2b | 10808 | -0.346 | -0.0015 | No |
| 75 | Erbb3 | 11133 | -0.393 | -0.0036 | No |
| 76 | Fdps | 11334 | -0.423 | 0.0062 | No |
| 77 | Smtn | 11360 | -0.428 | 0.0305 | No |
| 78 | Myl6b | 11452 | -0.442 | 0.0503 | No |
| 79 | Cdh13 | 11668 | -0.482 | 0.0625 | No |