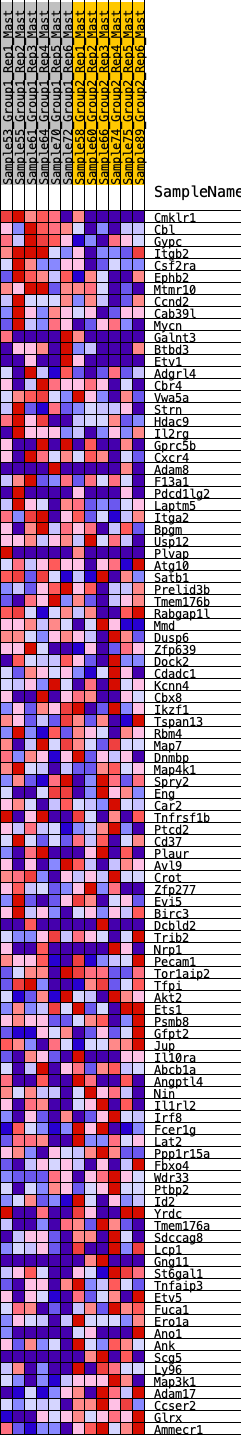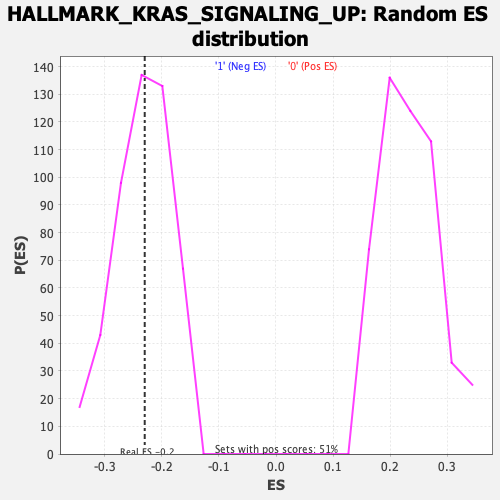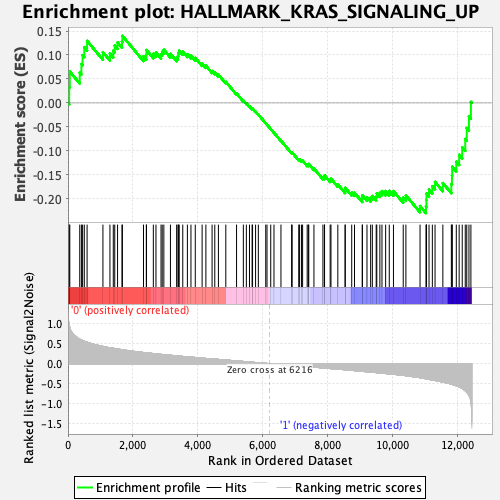
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | -0.22973114 |
| Normalized Enrichment Score (NES) | -0.9861958 |
| Nominal p-value | 0.47070706 |
| FDR q-value | 0.97726977 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cmklr1 | 37 | 0.979 | 0.0333 | No |
| 2 | Cbl | 55 | 0.902 | 0.0654 | No |
| 3 | Gypc | 362 | 0.610 | 0.0633 | No |
| 4 | Itgb2 | 413 | 0.593 | 0.0812 | No |
| 5 | Csf2ra | 450 | 0.577 | 0.0998 | No |
| 6 | Ephb2 | 502 | 0.560 | 0.1164 | No |
| 7 | Mtmr10 | 588 | 0.538 | 0.1295 | No |
| 8 | Ccnd2 | 1076 | 0.430 | 0.1060 | No |
| 9 | Cab39l | 1292 | 0.395 | 0.1032 | No |
| 10 | Mycn | 1395 | 0.383 | 0.1091 | No |
| 11 | Galnt3 | 1440 | 0.377 | 0.1196 | No |
| 12 | Btbd3 | 1527 | 0.364 | 0.1261 | No |
| 13 | Etv1 | 1663 | 0.347 | 0.1280 | No |
| 14 | Adgrl4 | 1676 | 0.346 | 0.1399 | No |
| 15 | Cbr4 | 2326 | 0.277 | 0.0976 | No |
| 16 | Vwa5a | 2414 | 0.270 | 0.1005 | No |
| 17 | Strn | 2417 | 0.269 | 0.1104 | No |
| 18 | Hdac9 | 2629 | 0.252 | 0.1026 | No |
| 19 | Il2rg | 2710 | 0.245 | 0.1052 | No |
| 20 | Gprc5b | 2866 | 0.231 | 0.1012 | No |
| 21 | Cxcr4 | 2902 | 0.227 | 0.1068 | No |
| 22 | Adam8 | 2953 | 0.224 | 0.1111 | No |
| 23 | F13a1 | 3156 | 0.208 | 0.1024 | No |
| 24 | Pdcd1lg2 | 3347 | 0.194 | 0.0942 | No |
| 25 | Laptm5 | 3390 | 0.191 | 0.0979 | No |
| 26 | Itga2 | 3401 | 0.190 | 0.1042 | No |
| 27 | Bpgm | 3426 | 0.188 | 0.1092 | No |
| 28 | Usp12 | 3536 | 0.180 | 0.1070 | No |
| 29 | Plvap | 3678 | 0.170 | 0.1019 | No |
| 30 | Atg10 | 3788 | 0.163 | 0.0991 | No |
| 31 | Satb1 | 3921 | 0.153 | 0.0941 | No |
| 32 | Prelid3b | 4132 | 0.139 | 0.0822 | No |
| 33 | Tmem176b | 4248 | 0.129 | 0.0777 | No |
| 34 | Rabgap1l | 4439 | 0.117 | 0.0667 | No |
| 35 | Mmd | 4522 | 0.112 | 0.0642 | No |
| 36 | Dusp6 | 4634 | 0.106 | 0.0591 | No |
| 37 | Zfp639 | 4862 | 0.091 | 0.0441 | No |
| 38 | Dock2 | 5193 | 0.068 | 0.0199 | No |
| 39 | Cdadc1 | 5402 | 0.054 | 0.0050 | No |
| 40 | Kcnn4 | 5495 | 0.047 | -0.0007 | No |
| 41 | Cbx8 | 5591 | 0.041 | -0.0068 | No |
| 42 | Ikzf1 | 5672 | 0.035 | -0.0120 | No |
| 43 | Tspan13 | 5686 | 0.034 | -0.0118 | No |
| 44 | Rbm4 | 5779 | 0.028 | -0.0182 | No |
| 45 | Map7 | 5864 | 0.023 | -0.0242 | No |
| 46 | Dnmbp | 6092 | 0.008 | -0.0423 | No |
| 47 | Map4k1 | 6131 | 0.005 | -0.0452 | No |
| 48 | Spry2 | 6246 | -0.001 | -0.0543 | No |
| 49 | Eng | 6346 | -0.009 | -0.0620 | No |
| 50 | Car2 | 6561 | -0.023 | -0.0785 | No |
| 51 | Tnfrsf1b | 6889 | -0.040 | -0.1036 | No |
| 52 | Ptcd2 | 6905 | -0.041 | -0.1033 | No |
| 53 | Cd37 | 7109 | -0.055 | -0.1177 | No |
| 54 | Plaur | 7135 | -0.057 | -0.1176 | No |
| 55 | Avl9 | 7195 | -0.061 | -0.1201 | No |
| 56 | Crot | 7228 | -0.063 | -0.1203 | No |
| 57 | Zfp277 | 7369 | -0.073 | -0.1289 | No |
| 58 | Evi5 | 7405 | -0.076 | -0.1290 | No |
| 59 | Birc3 | 7418 | -0.077 | -0.1271 | No |
| 60 | Dcbld2 | 7577 | -0.088 | -0.1366 | No |
| 61 | Trib2 | 7854 | -0.105 | -0.1551 | No |
| 62 | Nrp1 | 7901 | -0.108 | -0.1548 | No |
| 63 | Pecam1 | 7904 | -0.108 | -0.1510 | No |
| 64 | Tor1aip2 | 8077 | -0.122 | -0.1604 | No |
| 65 | Tfpi | 8102 | -0.123 | -0.1578 | No |
| 66 | Akt2 | 8313 | -0.136 | -0.1698 | No |
| 67 | Ets1 | 8538 | -0.151 | -0.1823 | No |
| 68 | Psmb8 | 8540 | -0.151 | -0.1768 | No |
| 69 | Gfpt2 | 8741 | -0.165 | -0.1868 | No |
| 70 | Jup | 8827 | -0.173 | -0.1873 | No |
| 71 | Il10ra | 9066 | -0.191 | -0.1995 | No |
| 72 | Abcb1a | 9070 | -0.192 | -0.1926 | No |
| 73 | Angptl4 | 9211 | -0.202 | -0.1965 | No |
| 74 | Nin | 9324 | -0.210 | -0.1978 | No |
| 75 | Il1rl2 | 9376 | -0.214 | -0.1940 | No |
| 76 | Irf8 | 9495 | -0.223 | -0.1953 | No |
| 77 | Fcer1g | 9514 | -0.225 | -0.1884 | No |
| 78 | Lat2 | 9608 | -0.232 | -0.1873 | No |
| 79 | Ppp1r15a | 9673 | -0.237 | -0.1837 | No |
| 80 | Fbxo4 | 9786 | -0.246 | -0.1836 | No |
| 81 | Wdr33 | 9898 | -0.254 | -0.1832 | No |
| 82 | Ptbp2 | 10027 | -0.266 | -0.1837 | No |
| 83 | Id2 | 10328 | -0.291 | -0.1972 | No |
| 84 | Yrdc | 10410 | -0.300 | -0.1926 | No |
| 85 | Tmem176a | 10846 | -0.350 | -0.2149 | No |
| 86 | Sdccag8 | 11030 | -0.377 | -0.2157 | Yes |
| 87 | Lcp1 | 11043 | -0.379 | -0.2026 | Yes |
| 88 | Gng11 | 11046 | -0.379 | -0.1887 | Yes |
| 89 | St6gal1 | 11123 | -0.391 | -0.1804 | Yes |
| 90 | Tnfaip3 | 11225 | -0.407 | -0.1734 | Yes |
| 91 | Etv5 | 11308 | -0.419 | -0.1645 | Yes |
| 92 | Fuca1 | 11549 | -0.459 | -0.1670 | Yes |
| 93 | Ero1a | 11810 | -0.512 | -0.1690 | Yes |
| 94 | Ano1 | 11830 | -0.516 | -0.1514 | Yes |
| 95 | Ank | 11837 | -0.517 | -0.1327 | Yes |
| 96 | Scg5 | 11963 | -0.550 | -0.1224 | Yes |
| 97 | Ly96 | 12051 | -0.579 | -0.1080 | Yes |
| 98 | Map3k1 | 12146 | -0.618 | -0.0926 | Yes |
| 99 | Adam17 | 12239 | -0.676 | -0.0750 | Yes |
| 100 | Ccser2 | 12285 | -0.715 | -0.0521 | Yes |
| 101 | Glrx | 12353 | -0.785 | -0.0284 | Yes |
| 102 | Ammecr1 | 12413 | -0.955 | 0.0023 | Yes |