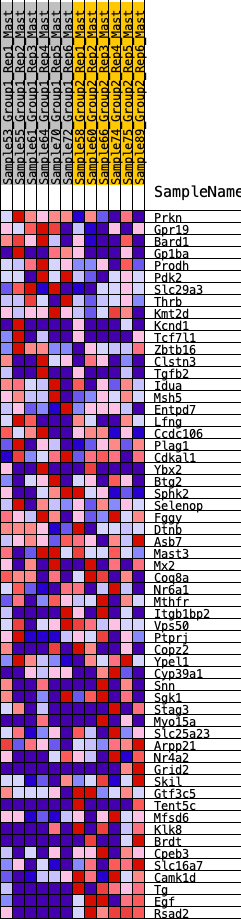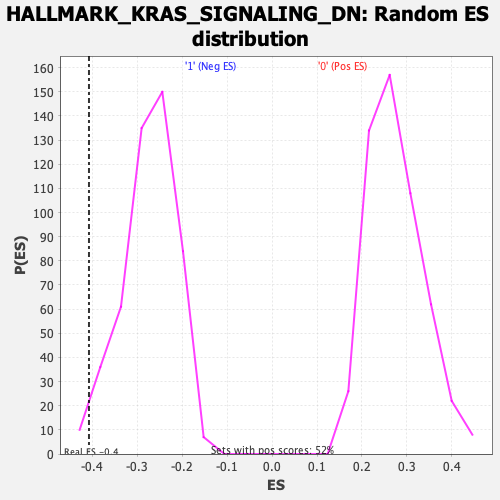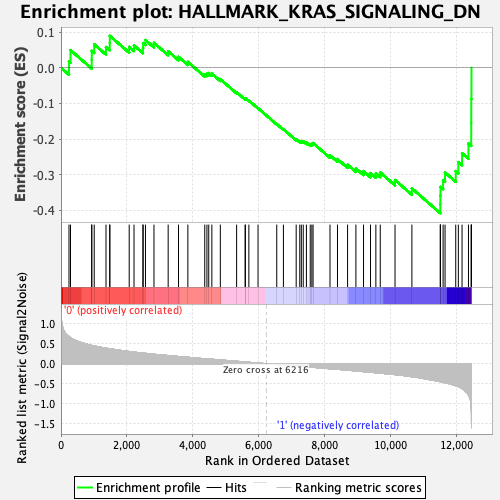
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.40781516 |
| Normalized Enrichment Score (NES) | -1.4900469 |
| Nominal p-value | 0.016563147 |
| FDR q-value | 0.22505635 |
| FWER p-Value | 0.328 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Prkn | 242 | 0.680 | 0.0176 | No |
| 2 | Gpr19 | 288 | 0.649 | 0.0494 | No |
| 3 | Bard1 | 931 | 0.456 | 0.0225 | No |
| 4 | Gp1ba | 933 | 0.455 | 0.0473 | No |
| 5 | Prodh | 1007 | 0.443 | 0.0656 | No |
| 6 | Pdk2 | 1364 | 0.387 | 0.0580 | No |
| 7 | Slc29a3 | 1478 | 0.372 | 0.0692 | No |
| 8 | Thrb | 1480 | 0.371 | 0.0894 | No |
| 9 | Kmt2d | 2067 | 0.302 | 0.0586 | No |
| 10 | Kcnd1 | 2213 | 0.289 | 0.0627 | No |
| 11 | Tcf7l1 | 2483 | 0.264 | 0.0554 | No |
| 12 | Zbtb16 | 2489 | 0.263 | 0.0693 | No |
| 13 | Clstn3 | 2560 | 0.257 | 0.0777 | No |
| 14 | Tgfb2 | 2818 | 0.235 | 0.0698 | No |
| 15 | Idua | 3248 | 0.202 | 0.0462 | No |
| 16 | Msh5 | 3561 | 0.178 | 0.0307 | No |
| 17 | Entpd7 | 3845 | 0.159 | 0.0165 | No |
| 18 | Lfng | 4357 | 0.123 | -0.0180 | No |
| 19 | Ccdc106 | 4423 | 0.119 | -0.0168 | No |
| 20 | Plag1 | 4478 | 0.115 | -0.0149 | No |
| 21 | Cdkal1 | 4572 | 0.109 | -0.0165 | No |
| 22 | Ybx2 | 4829 | 0.093 | -0.0321 | No |
| 23 | Btg2 | 5323 | 0.060 | -0.0686 | No |
| 24 | Sphk2 | 5580 | 0.042 | -0.0870 | No |
| 25 | Selenop | 5589 | 0.042 | -0.0854 | No |
| 26 | Fggy | 5694 | 0.033 | -0.0919 | No |
| 27 | Dtnb | 5972 | 0.017 | -0.1134 | No |
| 28 | Asb7 | 6539 | -0.021 | -0.1579 | No |
| 29 | Mast3 | 6741 | -0.032 | -0.1724 | No |
| 30 | Mx2 | 7128 | -0.057 | -0.2005 | No |
| 31 | Coq8a | 7237 | -0.064 | -0.2057 | No |
| 32 | Nr6a1 | 7284 | -0.068 | -0.2057 | No |
| 33 | Mthfr | 7342 | -0.071 | -0.2064 | No |
| 34 | Itgb1bp2 | 7438 | -0.078 | -0.2098 | No |
| 35 | Vps50 | 7554 | -0.086 | -0.2144 | No |
| 36 | Ptprj | 7603 | -0.090 | -0.2134 | No |
| 37 | Copz2 | 7640 | -0.092 | -0.2113 | No |
| 38 | Ypel1 | 8153 | -0.126 | -0.2457 | No |
| 39 | Cyp39a1 | 8380 | -0.141 | -0.2562 | No |
| 40 | Snn | 8686 | -0.161 | -0.2721 | No |
| 41 | Sgk1 | 8938 | -0.181 | -0.2824 | No |
| 42 | Stag3 | 9169 | -0.199 | -0.2901 | No |
| 43 | Myo15a | 9381 | -0.214 | -0.2955 | No |
| 44 | Slc25a23 | 9543 | -0.227 | -0.2961 | No |
| 45 | Arpp21 | 9676 | -0.237 | -0.2938 | No |
| 46 | Nr4a2 | 10124 | -0.273 | -0.3150 | No |
| 47 | Grid2 | 10635 | -0.325 | -0.3384 | No |
| 48 | Skil | 11496 | -0.449 | -0.3833 | Yes |
| 49 | Gtf3c5 | 11498 | -0.450 | -0.3588 | Yes |
| 50 | Tent5c | 11502 | -0.451 | -0.3344 | Yes |
| 51 | Mfsd6 | 11581 | -0.465 | -0.3153 | Yes |
| 52 | Klk8 | 11640 | -0.478 | -0.2938 | Yes |
| 53 | Brdt | 11964 | -0.551 | -0.2898 | Yes |
| 54 | Cpeb3 | 12043 | -0.575 | -0.2647 | Yes |
| 55 | Slc16a7 | 12155 | -0.621 | -0.2397 | Yes |
| 56 | Camk1d | 12350 | -0.783 | -0.2126 | Yes |
| 57 | Tg | 12434 | -1.187 | -0.1544 | Yes |
| 58 | Egf | 12435 | -1.223 | -0.0875 | Yes |
| 59 | Rsad2 | 12441 | -1.609 | 0.0000 | Yes |