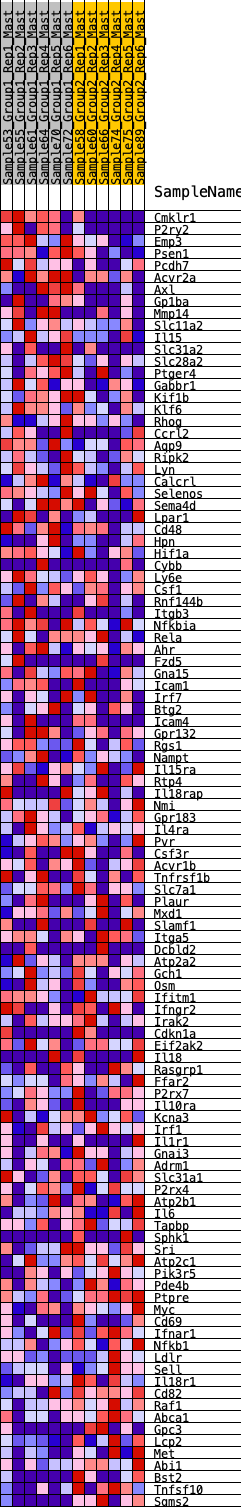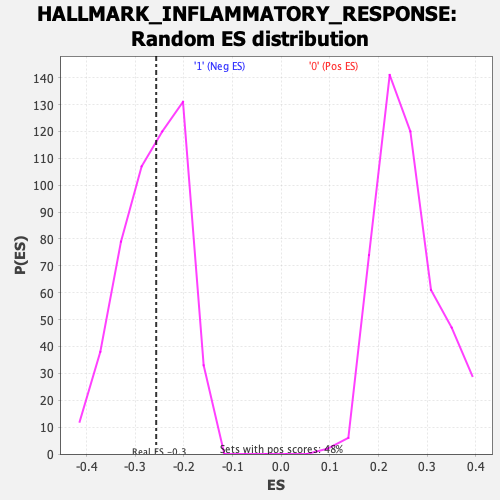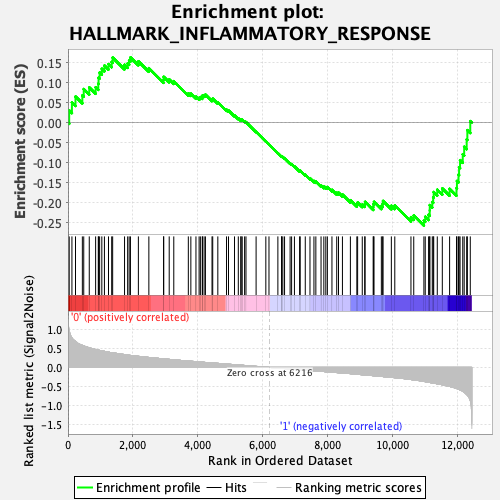
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.2570458 |
| Normalized Enrichment Score (NES) | -0.9742253 |
| Nominal p-value | 0.49807692 |
| FDR q-value | 0.88801605 |
| FWER p-Value | 0.992 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cmklr1 | 37 | 0.979 | 0.0305 | No |
| 2 | P2ry2 | 121 | 0.781 | 0.0505 | No |
| 3 | Emp3 | 231 | 0.688 | 0.0652 | No |
| 4 | Psen1 | 442 | 0.581 | 0.0680 | No |
| 5 | Pcdh7 | 485 | 0.564 | 0.0839 | No |
| 6 | Acvr2a | 655 | 0.518 | 0.0880 | No |
| 7 | Axl | 851 | 0.473 | 0.0883 | No |
| 8 | Gp1ba | 933 | 0.455 | 0.0973 | No |
| 9 | Mmp14 | 942 | 0.454 | 0.1122 | No |
| 10 | Slc11a2 | 974 | 0.448 | 0.1250 | No |
| 11 | Il15 | 1042 | 0.436 | 0.1345 | No |
| 12 | Slc31a2 | 1122 | 0.424 | 0.1426 | No |
| 13 | Slc28a2 | 1248 | 0.403 | 0.1462 | No |
| 14 | Ptger4 | 1346 | 0.389 | 0.1516 | No |
| 15 | Gabbr1 | 1376 | 0.385 | 0.1625 | No |
| 16 | Kif1b | 1742 | 0.339 | 0.1445 | No |
| 17 | Klf6 | 1838 | 0.327 | 0.1479 | No |
| 18 | Rhog | 1884 | 0.320 | 0.1552 | No |
| 19 | Ccrl2 | 1924 | 0.316 | 0.1629 | No |
| 20 | Aqp9 | 2168 | 0.292 | 0.1532 | No |
| 21 | Ripk2 | 2491 | 0.263 | 0.1360 | No |
| 22 | Lyn | 2944 | 0.224 | 0.1071 | No |
| 23 | Calcrl | 2949 | 0.224 | 0.1144 | No |
| 24 | Selenos | 3118 | 0.212 | 0.1080 | No |
| 25 | Sema4d | 3259 | 0.201 | 0.1035 | No |
| 26 | Lpar1 | 3707 | 0.169 | 0.0731 | No |
| 27 | Cd48 | 3785 | 0.163 | 0.0724 | No |
| 28 | Hpn | 3942 | 0.151 | 0.0649 | No |
| 29 | Hif1a | 4039 | 0.144 | 0.0621 | No |
| 30 | Cybb | 4078 | 0.143 | 0.0639 | No |
| 31 | Ly6e | 4136 | 0.138 | 0.0640 | No |
| 32 | Csf1 | 4145 | 0.138 | 0.0680 | No |
| 33 | Rnf144b | 4193 | 0.134 | 0.0688 | No |
| 34 | Itgb3 | 4234 | 0.130 | 0.0700 | No |
| 35 | Nfkbia | 4437 | 0.118 | 0.0577 | No |
| 36 | Rela | 4459 | 0.116 | 0.0599 | No |
| 37 | Ahr | 4616 | 0.107 | 0.0509 | No |
| 38 | Fzd5 | 4884 | 0.090 | 0.0324 | No |
| 39 | Gna15 | 4945 | 0.084 | 0.0304 | No |
| 40 | Icam1 | 5130 | 0.072 | 0.0180 | No |
| 41 | Irf7 | 5247 | 0.065 | 0.0108 | No |
| 42 | Btg2 | 5323 | 0.060 | 0.0067 | No |
| 43 | Icam4 | 5334 | 0.059 | 0.0079 | No |
| 44 | Gpr132 | 5367 | 0.056 | 0.0073 | No |
| 45 | Rgs1 | 5441 | 0.051 | 0.0031 | No |
| 46 | Nampt | 5483 | 0.048 | 0.0014 | No |
| 47 | Il15ra | 5795 | 0.027 | -0.0229 | No |
| 48 | Rtp4 | 6095 | 0.008 | -0.0468 | No |
| 49 | Il18rap | 6192 | 0.002 | -0.0546 | No |
| 50 | Nmi | 6466 | -0.015 | -0.0762 | No |
| 51 | Gpr183 | 6579 | -0.024 | -0.0845 | No |
| 52 | Il4ra | 6585 | -0.024 | -0.0840 | No |
| 53 | Pvr | 6619 | -0.026 | -0.0858 | No |
| 54 | Csf3r | 6672 | -0.028 | -0.0891 | No |
| 55 | Acvr1b | 6843 | -0.037 | -0.1016 | No |
| 56 | Tnfrsf1b | 6889 | -0.040 | -0.1039 | No |
| 57 | Slc7a1 | 6976 | -0.047 | -0.1093 | No |
| 58 | Plaur | 7135 | -0.057 | -0.1201 | No |
| 59 | Mxd1 | 7154 | -0.058 | -0.1196 | No |
| 60 | Slamf1 | 7310 | -0.069 | -0.1298 | No |
| 61 | Itga5 | 7455 | -0.080 | -0.1387 | No |
| 62 | Dcbld2 | 7577 | -0.088 | -0.1455 | No |
| 63 | Atp2a2 | 7636 | -0.092 | -0.1471 | No |
| 64 | Gch1 | 7796 | -0.100 | -0.1566 | No |
| 65 | Osm | 7882 | -0.107 | -0.1598 | No |
| 66 | Ifitm1 | 7943 | -0.112 | -0.1609 | No |
| 67 | Ifngr2 | 7991 | -0.115 | -0.1607 | No |
| 68 | Irak2 | 8132 | -0.125 | -0.1678 | No |
| 69 | Cdkn1a | 8279 | -0.134 | -0.1751 | No |
| 70 | Eif2ak2 | 8334 | -0.137 | -0.1747 | No |
| 71 | Il18 | 8455 | -0.146 | -0.1795 | No |
| 72 | Rasgrp1 | 8700 | -0.162 | -0.1937 | No |
| 73 | Ffar2 | 8899 | -0.178 | -0.2037 | No |
| 74 | P2rx7 | 8923 | -0.180 | -0.1994 | No |
| 75 | Il10ra | 9066 | -0.191 | -0.2043 | No |
| 76 | Kcna3 | 9142 | -0.197 | -0.2037 | No |
| 77 | Irf1 | 9152 | -0.197 | -0.1977 | No |
| 78 | Il1r1 | 9405 | -0.216 | -0.2107 | No |
| 79 | Gnai3 | 9408 | -0.216 | -0.2035 | No |
| 80 | Adrm1 | 9428 | -0.217 | -0.1976 | No |
| 81 | Slc31a1 | 9654 | -0.235 | -0.2078 | No |
| 82 | P2rx4 | 9694 | -0.238 | -0.2028 | No |
| 83 | Atp2b1 | 9705 | -0.239 | -0.1955 | No |
| 84 | Il6 | 9962 | -0.260 | -0.2073 | No |
| 85 | Tapbp | 10068 | -0.268 | -0.2066 | No |
| 86 | Sphk1 | 10566 | -0.318 | -0.2361 | No |
| 87 | Sri | 10652 | -0.327 | -0.2317 | No |
| 88 | Atp2c1 | 10965 | -0.367 | -0.2445 | Yes |
| 89 | Pik3r5 | 11006 | -0.374 | -0.2350 | Yes |
| 90 | Pde4b | 11109 | -0.389 | -0.2299 | Yes |
| 91 | Ptpre | 11144 | -0.394 | -0.2192 | Yes |
| 92 | Myc | 11146 | -0.395 | -0.2058 | Yes |
| 93 | Cd69 | 11223 | -0.407 | -0.1980 | Yes |
| 94 | Ifnar1 | 11252 | -0.411 | -0.1862 | Yes |
| 95 | Nfkb1 | 11267 | -0.413 | -0.1732 | Yes |
| 96 | Ldlr | 11377 | -0.431 | -0.1673 | Yes |
| 97 | Sell | 11532 | -0.456 | -0.1642 | Yes |
| 98 | Il18r1 | 11757 | -0.498 | -0.1653 | Yes |
| 99 | Cd82 | 11969 | -0.552 | -0.1636 | Yes |
| 100 | Raf1 | 11983 | -0.558 | -0.1455 | Yes |
| 101 | Abca1 | 12032 | -0.570 | -0.1299 | Yes |
| 102 | Gpc3 | 12049 | -0.578 | -0.1114 | Yes |
| 103 | Lcp2 | 12079 | -0.589 | -0.0936 | Yes |
| 104 | Met | 12162 | -0.624 | -0.0789 | Yes |
| 105 | Abi1 | 12206 | -0.651 | -0.0602 | Yes |
| 106 | Bst2 | 12283 | -0.713 | -0.0419 | Yes |
| 107 | Tnfsf10 | 12304 | -0.726 | -0.0187 | Yes |
| 108 | Sgms2 | 12394 | -0.870 | 0.0038 | Yes |