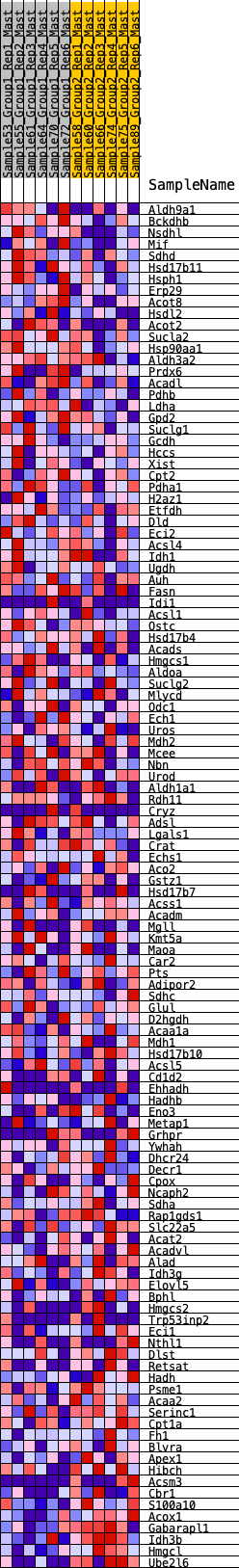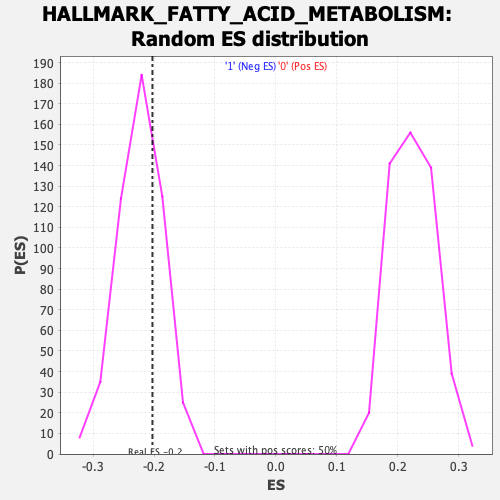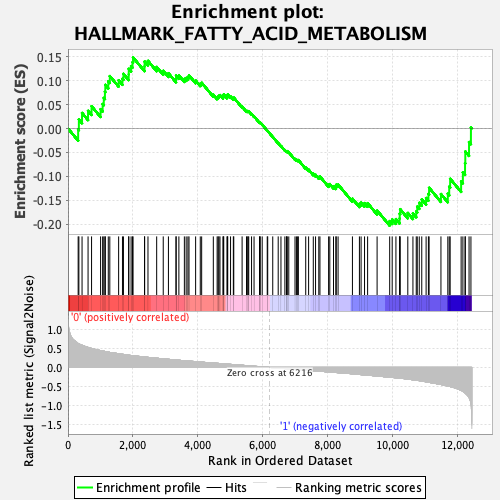
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.20255683 |
| Normalized Enrichment Score (NES) | -0.91001457 |
| Nominal p-value | 0.7025948 |
| FDR q-value | 0.84210086 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Aldh9a1 | 313 | 0.630 | -0.0023 | No |
| 2 | Bckdhb | 335 | 0.620 | 0.0188 | No |
| 3 | Nsdhl | 433 | 0.585 | 0.0324 | No |
| 4 | Mif | 618 | 0.528 | 0.0368 | No |
| 5 | Sdhd | 726 | 0.499 | 0.0464 | No |
| 6 | Hsd17b11 | 1004 | 0.443 | 0.0402 | No |
| 7 | Hsph1 | 1067 | 0.432 | 0.0511 | No |
| 8 | Erp29 | 1103 | 0.426 | 0.0639 | No |
| 9 | Acot8 | 1138 | 0.420 | 0.0766 | No |
| 10 | Hsdl2 | 1150 | 0.418 | 0.0910 | No |
| 11 | Acot2 | 1241 | 0.403 | 0.0985 | No |
| 12 | Sucla2 | 1288 | 0.395 | 0.1093 | No |
| 13 | Hsp90aa1 | 1560 | 0.361 | 0.1006 | No |
| 14 | Aldh3a2 | 1678 | 0.346 | 0.1037 | No |
| 15 | Prdx6 | 1708 | 0.343 | 0.1140 | No |
| 16 | Acadl | 1870 | 0.322 | 0.1127 | No |
| 17 | Pdhb | 1872 | 0.322 | 0.1245 | No |
| 18 | Ldha | 1944 | 0.314 | 0.1303 | No |
| 19 | Gpd2 | 1988 | 0.310 | 0.1381 | No |
| 20 | Suclg1 | 2004 | 0.308 | 0.1482 | No |
| 21 | Gcdh | 2358 | 0.275 | 0.1297 | No |
| 22 | Hccs | 2361 | 0.275 | 0.1396 | No |
| 23 | Xist | 2464 | 0.265 | 0.1411 | No |
| 24 | Cpt2 | 2732 | 0.243 | 0.1283 | No |
| 25 | Pdha1 | 2934 | 0.225 | 0.1203 | No |
| 26 | H2az1 | 3094 | 0.214 | 0.1152 | No |
| 27 | Etfdh | 3329 | 0.195 | 0.1034 | No |
| 28 | Dld | 3330 | 0.195 | 0.1106 | No |
| 29 | Eci2 | 3416 | 0.189 | 0.1106 | No |
| 30 | Acsl4 | 3585 | 0.176 | 0.1035 | No |
| 31 | Idh1 | 3640 | 0.172 | 0.1054 | No |
| 32 | Ugdh | 3694 | 0.169 | 0.1073 | No |
| 33 | Auh | 3728 | 0.167 | 0.1108 | No |
| 34 | Fasn | 3932 | 0.152 | 0.0999 | No |
| 35 | Idi1 | 4075 | 0.143 | 0.0936 | No |
| 36 | Acsl1 | 4118 | 0.140 | 0.0953 | No |
| 37 | Ostc | 4477 | 0.115 | 0.0705 | No |
| 38 | Hsd17b4 | 4595 | 0.107 | 0.0649 | No |
| 39 | Acads | 4625 | 0.106 | 0.0665 | No |
| 40 | Hmgcs1 | 4649 | 0.105 | 0.0685 | No |
| 41 | Aldoa | 4682 | 0.102 | 0.0696 | No |
| 42 | Suclg2 | 4775 | 0.096 | 0.0657 | No |
| 43 | Mlycd | 4787 | 0.095 | 0.0683 | No |
| 44 | Odc1 | 4798 | 0.094 | 0.0710 | No |
| 45 | Ech1 | 4899 | 0.088 | 0.0661 | No |
| 46 | Uros | 4912 | 0.087 | 0.0683 | No |
| 47 | Mdh2 | 4921 | 0.086 | 0.0708 | No |
| 48 | Mcee | 5008 | 0.080 | 0.0668 | No |
| 49 | Nbn | 5094 | 0.074 | 0.0626 | No |
| 50 | Urod | 5107 | 0.073 | 0.0643 | No |
| 51 | Aldh1a1 | 5364 | 0.056 | 0.0456 | No |
| 52 | Rdh11 | 5500 | 0.047 | 0.0364 | No |
| 53 | Cryz | 5521 | 0.046 | 0.0364 | No |
| 54 | Adsl | 5548 | 0.044 | 0.0359 | No |
| 55 | Lgals1 | 5568 | 0.043 | 0.0360 | No |
| 56 | Crat | 5655 | 0.036 | 0.0303 | No |
| 57 | Echs1 | 5732 | 0.031 | 0.0253 | No |
| 58 | Aco2 | 5900 | 0.021 | 0.0125 | No |
| 59 | Gstz1 | 5913 | 0.020 | 0.0122 | No |
| 60 | Hsd17b7 | 5920 | 0.019 | 0.0125 | No |
| 61 | Acss1 | 5983 | 0.016 | 0.0080 | No |
| 62 | Acadm | 6138 | 0.005 | -0.0043 | No |
| 63 | Mgll | 6153 | 0.004 | -0.0053 | No |
| 64 | Kmt5a | 6308 | -0.007 | -0.0176 | No |
| 65 | Maoa | 6476 | -0.016 | -0.0305 | No |
| 66 | Car2 | 6561 | -0.023 | -0.0365 | No |
| 67 | Pts | 6682 | -0.029 | -0.0452 | No |
| 68 | Adipor2 | 6731 | -0.031 | -0.0479 | No |
| 69 | Sdhc | 6739 | -0.032 | -0.0473 | No |
| 70 | Glul | 6755 | -0.033 | -0.0473 | No |
| 71 | D2hgdh | 6802 | -0.035 | -0.0498 | No |
| 72 | Acaa1a | 6994 | -0.048 | -0.0635 | No |
| 73 | Mdh1 | 7038 | -0.051 | -0.0652 | No |
| 74 | Hsd17b10 | 7074 | -0.053 | -0.0661 | No |
| 75 | Acsl5 | 7097 | -0.055 | -0.0659 | No |
| 76 | Cd1d2 | 7324 | -0.070 | -0.0816 | No |
| 77 | Ehhadh | 7413 | -0.077 | -0.0859 | No |
| 78 | Hadhb | 7557 | -0.086 | -0.0944 | No |
| 79 | Eno3 | 7627 | -0.091 | -0.0966 | No |
| 80 | Metap1 | 7727 | -0.097 | -0.1011 | No |
| 81 | Grhpr | 7762 | -0.099 | -0.1002 | No |
| 82 | Ywhah | 8029 | -0.118 | -0.1175 | No |
| 83 | Dhcr24 | 8066 | -0.120 | -0.1160 | No |
| 84 | Decr1 | 8180 | -0.128 | -0.1204 | No |
| 85 | Cpox | 8253 | -0.132 | -0.1214 | No |
| 86 | Ncaph2 | 8263 | -0.133 | -0.1173 | No |
| 87 | Sdha | 8321 | -0.136 | -0.1169 | No |
| 88 | Rap1gds1 | 8763 | -0.168 | -0.1465 | No |
| 89 | Slc22a5 | 8978 | -0.184 | -0.1571 | No |
| 90 | Acat2 | 9030 | -0.189 | -0.1544 | No |
| 91 | Acadvl | 9136 | -0.196 | -0.1557 | No |
| 92 | Alad | 9231 | -0.203 | -0.1558 | No |
| 93 | Idh3g | 9523 | -0.225 | -0.1712 | No |
| 94 | Elovl5 | 9911 | -0.255 | -0.1932 | Yes |
| 95 | Bphl | 9988 | -0.262 | -0.1897 | Yes |
| 96 | Hmgcs2 | 10103 | -0.271 | -0.1890 | Yes |
| 97 | Trp53inp2 | 10216 | -0.281 | -0.1878 | Yes |
| 98 | Eci1 | 10220 | -0.281 | -0.1777 | Yes |
| 99 | Nthl1 | 10234 | -0.283 | -0.1684 | Yes |
| 100 | Dlst | 10469 | -0.305 | -0.1762 | Yes |
| 101 | Retsat | 10627 | -0.325 | -0.1770 | Yes |
| 102 | Hadh | 10729 | -0.335 | -0.1729 | Yes |
| 103 | Psme1 | 10760 | -0.339 | -0.1629 | Yes |
| 104 | Acaa2 | 10822 | -0.347 | -0.1551 | Yes |
| 105 | Serinc1 | 10899 | -0.359 | -0.1481 | Yes |
| 106 | Cpt1a | 11033 | -0.378 | -0.1450 | Yes |
| 107 | Fh1 | 11104 | -0.388 | -0.1364 | Yes |
| 108 | Blvra | 11125 | -0.391 | -0.1237 | Yes |
| 109 | Apex1 | 11490 | -0.448 | -0.1368 | Yes |
| 110 | Hibch | 11702 | -0.487 | -0.1360 | Yes |
| 111 | Acsm3 | 11745 | -0.496 | -0.1212 | Yes |
| 112 | Cbr1 | 11774 | -0.503 | -0.1050 | Yes |
| 113 | S100a10 | 12112 | -0.600 | -0.1103 | Yes |
| 114 | Acox1 | 12166 | -0.628 | -0.0915 | Yes |
| 115 | Gabarapl1 | 12233 | -0.669 | -0.0723 | Yes |
| 116 | Idh3b | 12241 | -0.677 | -0.0480 | Yes |
| 117 | Hmgcl | 12356 | -0.790 | -0.0282 | Yes |
| 118 | Ube2l6 | 12414 | -0.955 | 0.0022 | Yes |