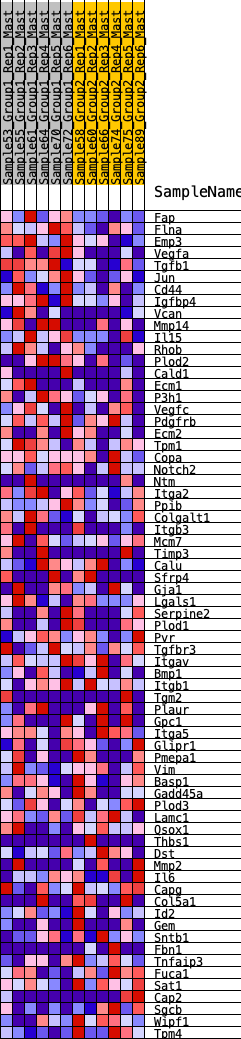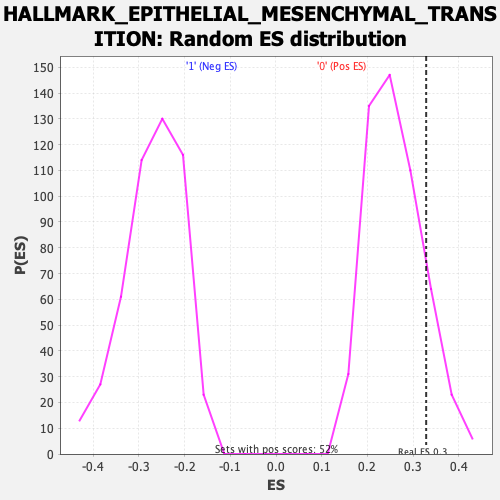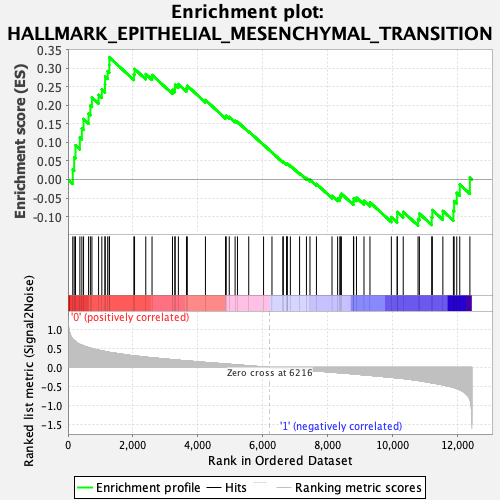
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.3288081 |
| Normalized Enrichment Score (NES) | 1.255105 |
| Nominal p-value | 0.13953489 |
| FDR q-value | 0.71623564 |
| FWER p-Value | 0.81 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fap | 147 | 0.749 | 0.0267 | Yes |
| 2 | Flna | 191 | 0.713 | 0.0599 | Yes |
| 3 | Emp3 | 231 | 0.688 | 0.0921 | Yes |
| 4 | Vegfa | 366 | 0.607 | 0.1125 | Yes |
| 5 | Tgfb1 | 427 | 0.586 | 0.1378 | Yes |
| 6 | Jun | 479 | 0.566 | 0.1629 | Yes |
| 7 | Cd44 | 634 | 0.522 | 0.1773 | Yes |
| 8 | Igfbp4 | 689 | 0.510 | 0.1992 | Yes |
| 9 | Vcan | 737 | 0.497 | 0.2209 | Yes |
| 10 | Mmp14 | 942 | 0.454 | 0.2278 | Yes |
| 11 | Il15 | 1042 | 0.436 | 0.2422 | Yes |
| 12 | Rhob | 1139 | 0.420 | 0.2561 | Yes |
| 13 | Plod2 | 1144 | 0.419 | 0.2773 | Yes |
| 14 | Cald1 | 1223 | 0.405 | 0.2919 | Yes |
| 15 | Ecm1 | 1273 | 0.399 | 0.3084 | Yes |
| 16 | P3h1 | 1275 | 0.398 | 0.3288 | Yes |
| 17 | Vegfc | 2030 | 0.306 | 0.2836 | No |
| 18 | Pdgfrb | 2050 | 0.305 | 0.2978 | No |
| 19 | Ecm2 | 2396 | 0.271 | 0.2838 | No |
| 20 | Tpm1 | 2589 | 0.255 | 0.2814 | No |
| 21 | Copa | 3219 | 0.204 | 0.2411 | No |
| 22 | Notch2 | 3293 | 0.199 | 0.2454 | No |
| 23 | Ntm | 3300 | 0.198 | 0.2551 | No |
| 24 | Itga2 | 3401 | 0.190 | 0.2568 | No |
| 25 | Ppib | 3650 | 0.172 | 0.2456 | No |
| 26 | Colgalt1 | 3675 | 0.170 | 0.2524 | No |
| 27 | Itgb3 | 4234 | 0.130 | 0.2140 | No |
| 28 | Mcm7 | 4853 | 0.092 | 0.1688 | No |
| 29 | Timp3 | 4875 | 0.090 | 0.1718 | No |
| 30 | Calu | 4968 | 0.083 | 0.1686 | No |
| 31 | Sfrp4 | 5148 | 0.071 | 0.1578 | No |
| 32 | Gja1 | 5221 | 0.066 | 0.1554 | No |
| 33 | Lgals1 | 5568 | 0.043 | 0.1296 | No |
| 34 | Serpine2 | 6025 | 0.013 | 0.0934 | No |
| 35 | Plod1 | 6284 | -0.005 | 0.0728 | No |
| 36 | Pvr | 6619 | -0.026 | 0.0471 | No |
| 37 | Tgfbr3 | 6633 | -0.027 | 0.0474 | No |
| 38 | Itgav | 6734 | -0.032 | 0.0410 | No |
| 39 | Bmp1 | 6751 | -0.033 | 0.0414 | No |
| 40 | Itgb1 | 6753 | -0.033 | 0.0430 | No |
| 41 | Tgm2 | 6851 | -0.038 | 0.0371 | No |
| 42 | Plaur | 7135 | -0.057 | 0.0171 | No |
| 43 | Gpc1 | 7346 | -0.072 | 0.0039 | No |
| 44 | Itga5 | 7455 | -0.080 | -0.0008 | No |
| 45 | Glipr1 | 7654 | -0.092 | -0.0120 | No |
| 46 | Pmepa1 | 8134 | -0.125 | -0.0443 | No |
| 47 | Vim | 8307 | -0.135 | -0.0512 | No |
| 48 | Basp1 | 8376 | -0.141 | -0.0495 | No |
| 49 | Gadd45a | 8391 | -0.142 | -0.0433 | No |
| 50 | Plod3 | 8423 | -0.144 | -0.0384 | No |
| 51 | Lamc1 | 8795 | -0.171 | -0.0596 | No |
| 52 | Qsox1 | 8803 | -0.171 | -0.0514 | No |
| 53 | Thbs1 | 8887 | -0.178 | -0.0489 | No |
| 54 | Dst | 9119 | -0.195 | -0.0575 | No |
| 55 | Mmp2 | 9303 | -0.208 | -0.0616 | No |
| 56 | Il6 | 9962 | -0.260 | -0.1014 | No |
| 57 | Capg | 10142 | -0.275 | -0.1017 | No |
| 58 | Col5a1 | 10144 | -0.275 | -0.0876 | No |
| 59 | Id2 | 10328 | -0.291 | -0.0874 | No |
| 60 | Gem | 10785 | -0.343 | -0.1067 | No |
| 61 | Sntb1 | 10826 | -0.347 | -0.0920 | No |
| 62 | Fbn1 | 11205 | -0.404 | -0.1018 | No |
| 63 | Tnfaip3 | 11225 | -0.407 | -0.0824 | No |
| 64 | Fuca1 | 11549 | -0.459 | -0.0849 | No |
| 65 | Sat1 | 11874 | -0.526 | -0.0840 | No |
| 66 | Cap2 | 11897 | -0.532 | -0.0584 | No |
| 67 | Sgcb | 11977 | -0.556 | -0.0362 | No |
| 68 | Wipf1 | 12070 | -0.586 | -0.0135 | No |
| 69 | Tpm4 | 12382 | -0.844 | 0.0048 | No |