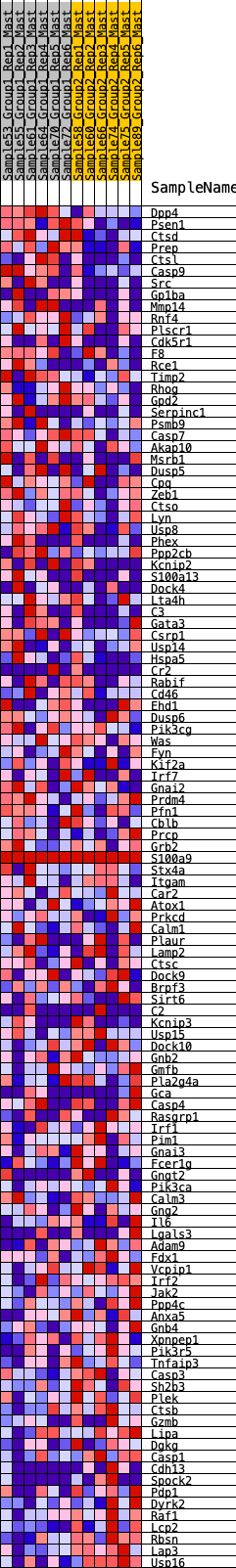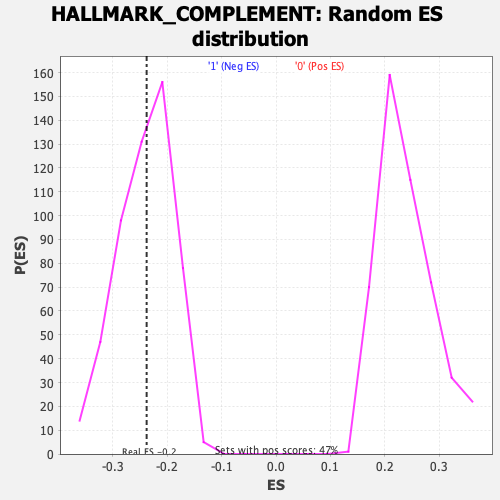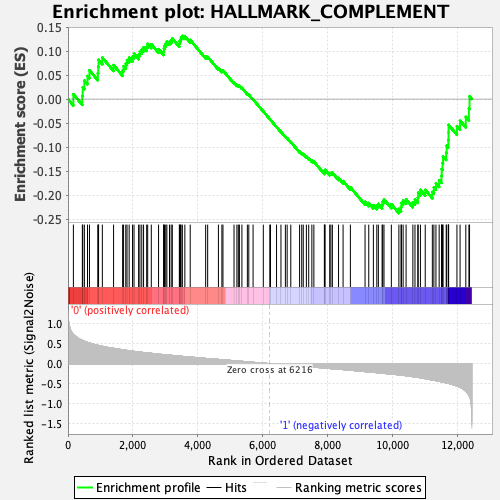
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.2375495 |
| Normalized Enrichment Score (NES) | -0.99493665 |
| Nominal p-value | 0.45746693 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dpp4 | 166 | 0.729 | 0.0101 | No |
| 2 | Psen1 | 442 | 0.581 | 0.0067 | No |
| 3 | Ctsd | 454 | 0.576 | 0.0245 | No |
| 4 | Prep | 504 | 0.558 | 0.0386 | No |
| 5 | Ctsl | 601 | 0.533 | 0.0480 | No |
| 6 | Casp9 | 661 | 0.517 | 0.0600 | No |
| 7 | Src | 920 | 0.457 | 0.0539 | No |
| 8 | Gp1ba | 933 | 0.455 | 0.0677 | No |
| 9 | Mmp14 | 942 | 0.454 | 0.0818 | No |
| 10 | Rnf4 | 1058 | 0.433 | 0.0865 | No |
| 11 | Plscr1 | 1404 | 0.382 | 0.0709 | No |
| 12 | Cdk5r1 | 1683 | 0.345 | 0.0595 | No |
| 13 | F8 | 1712 | 0.342 | 0.0683 | No |
| 14 | Rce1 | 1780 | 0.334 | 0.0737 | No |
| 15 | Timp2 | 1818 | 0.330 | 0.0814 | No |
| 16 | Rhog | 1884 | 0.320 | 0.0865 | No |
| 17 | Gpd2 | 1988 | 0.310 | 0.0882 | No |
| 18 | Serpinc1 | 2034 | 0.306 | 0.0944 | No |
| 19 | Psmb9 | 2175 | 0.291 | 0.0925 | No |
| 20 | Casp7 | 2216 | 0.288 | 0.0986 | No |
| 21 | Akap10 | 2273 | 0.282 | 0.1032 | No |
| 22 | Msrb1 | 2328 | 0.277 | 0.1078 | No |
| 23 | Dusp5 | 2424 | 0.269 | 0.1088 | No |
| 24 | Cpq | 2455 | 0.267 | 0.1150 | No |
| 25 | Zeb1 | 2566 | 0.256 | 0.1144 | No |
| 26 | Ctso | 2790 | 0.238 | 0.1040 | No |
| 27 | Lyn | 2944 | 0.224 | 0.0989 | No |
| 28 | Usp8 | 2957 | 0.223 | 0.1051 | No |
| 29 | Phex | 2974 | 0.222 | 0.1110 | No |
| 30 | Ppp2cb | 3012 | 0.219 | 0.1151 | No |
| 31 | Kcnip2 | 3046 | 0.217 | 0.1195 | No |
| 32 | S100a13 | 3127 | 0.211 | 0.1199 | No |
| 33 | Dock4 | 3178 | 0.207 | 0.1225 | No |
| 34 | Lta4h | 3214 | 0.205 | 0.1263 | No |
| 35 | C3 | 3430 | 0.188 | 0.1150 | No |
| 36 | Gata3 | 3436 | 0.188 | 0.1206 | No |
| 37 | Csrp1 | 3467 | 0.184 | 0.1242 | No |
| 38 | Usp14 | 3483 | 0.183 | 0.1289 | No |
| 39 | Hspa5 | 3523 | 0.181 | 0.1316 | No |
| 40 | Cr2 | 3600 | 0.175 | 0.1311 | No |
| 41 | Rabif | 3765 | 0.164 | 0.1231 | No |
| 42 | Cd46 | 4241 | 0.130 | 0.0888 | No |
| 43 | Ehd1 | 4303 | 0.126 | 0.0879 | No |
| 44 | Dusp6 | 4634 | 0.106 | 0.0646 | No |
| 45 | Pik3cg | 4741 | 0.098 | 0.0592 | No |
| 46 | Was | 4774 | 0.096 | 0.0597 | No |
| 47 | Fyn | 5115 | 0.073 | 0.0345 | No |
| 48 | Kif2a | 5204 | 0.067 | 0.0295 | No |
| 49 | Irf7 | 5247 | 0.065 | 0.0282 | No |
| 50 | Gnai2 | 5281 | 0.062 | 0.0275 | No |
| 51 | Prdm4 | 5358 | 0.057 | 0.0232 | No |
| 52 | Pfn1 | 5526 | 0.045 | 0.0111 | No |
| 53 | Cblb | 5570 | 0.043 | 0.0090 | No |
| 54 | Prcp | 5702 | 0.033 | -0.0005 | No |
| 55 | Grb2 | 6017 | 0.013 | -0.0256 | No |
| 56 | S100a9 | 6218 | 0.000 | -0.0418 | No |
| 57 | Stx4a | 6238 | -0.001 | -0.0433 | No |
| 58 | Itgam | 6425 | -0.012 | -0.0580 | No |
| 59 | Car2 | 6561 | -0.023 | -0.0682 | No |
| 60 | Atox1 | 6697 | -0.030 | -0.0782 | No |
| 61 | Prkcd | 6752 | -0.033 | -0.0815 | No |
| 62 | Calm1 | 6864 | -0.038 | -0.0893 | No |
| 63 | Plaur | 7135 | -0.057 | -0.1093 | No |
| 64 | Lamp2 | 7201 | -0.062 | -0.1126 | No |
| 65 | Ctsc | 7248 | -0.065 | -0.1142 | No |
| 66 | Dock9 | 7344 | -0.072 | -0.1196 | No |
| 67 | Brpf3 | 7416 | -0.077 | -0.1229 | No |
| 68 | Sirt6 | 7513 | -0.084 | -0.1280 | No |
| 69 | C2 | 7575 | -0.088 | -0.1301 | No |
| 70 | Kcnip3 | 7898 | -0.108 | -0.1527 | No |
| 71 | Usp15 | 7905 | -0.108 | -0.1497 | No |
| 72 | Dock10 | 7920 | -0.110 | -0.1472 | No |
| 73 | Gnb2 | 8063 | -0.120 | -0.1549 | No |
| 74 | Gmfb | 8099 | -0.123 | -0.1537 | No |
| 75 | Pla2g4a | 8145 | -0.126 | -0.1533 | No |
| 76 | Gca | 8335 | -0.137 | -0.1642 | No |
| 77 | Casp4 | 8475 | -0.147 | -0.1707 | No |
| 78 | Rasgrp1 | 8700 | -0.162 | -0.1836 | No |
| 79 | Irf1 | 9152 | -0.197 | -0.2138 | No |
| 80 | Pim1 | 9266 | -0.206 | -0.2163 | No |
| 81 | Gnai3 | 9408 | -0.216 | -0.2208 | No |
| 82 | Fcer1g | 9514 | -0.225 | -0.2220 | No |
| 83 | Gngt2 | 9562 | -0.229 | -0.2184 | No |
| 84 | Pik3ca | 9677 | -0.237 | -0.2199 | No |
| 85 | Calm3 | 9696 | -0.238 | -0.2137 | No |
| 86 | Gng2 | 9735 | -0.243 | -0.2089 | No |
| 87 | Il6 | 9962 | -0.260 | -0.2188 | No |
| 88 | Lgals3 | 10194 | -0.279 | -0.2285 | Yes |
| 89 | Adam9 | 10261 | -0.285 | -0.2246 | Yes |
| 90 | Fdx1 | 10272 | -0.287 | -0.2161 | Yes |
| 91 | Vcpip1 | 10325 | -0.291 | -0.2109 | Yes |
| 92 | Irf2 | 10416 | -0.300 | -0.2085 | Yes |
| 93 | Jak2 | 10625 | -0.325 | -0.2148 | Yes |
| 94 | Ppp4c | 10690 | -0.331 | -0.2093 | Yes |
| 95 | Anxa5 | 10778 | -0.341 | -0.2053 | Yes |
| 96 | Gnb4 | 10788 | -0.343 | -0.1949 | Yes |
| 97 | Xpnpep1 | 10858 | -0.352 | -0.1891 | Yes |
| 98 | Pik3r5 | 11006 | -0.374 | -0.1889 | Yes |
| 99 | Tnfaip3 | 11225 | -0.407 | -0.1934 | Yes |
| 100 | Casp3 | 11272 | -0.413 | -0.1838 | Yes |
| 101 | Sh2b3 | 11336 | -0.424 | -0.1751 | Yes |
| 102 | Plek | 11435 | -0.440 | -0.1688 | Yes |
| 103 | Ctsb | 11507 | -0.451 | -0.1600 | Yes |
| 104 | Gzmb | 11514 | -0.453 | -0.1458 | Yes |
| 105 | Lipa | 11540 | -0.458 | -0.1330 | Yes |
| 106 | Dgkg | 11555 | -0.460 | -0.1192 | Yes |
| 107 | Casp1 | 11652 | -0.480 | -0.1114 | Yes |
| 108 | Cdh13 | 11668 | -0.482 | -0.0970 | Yes |
| 109 | Spock2 | 11719 | -0.491 | -0.0851 | Yes |
| 110 | Pdp1 | 11724 | -0.492 | -0.0695 | Yes |
| 111 | Dyrk2 | 11726 | -0.492 | -0.0537 | Yes |
| 112 | Raf1 | 11983 | -0.558 | -0.0563 | Yes |
| 113 | Lcp2 | 12079 | -0.589 | -0.0450 | Yes |
| 114 | Rbsn | 12256 | -0.684 | -0.0371 | Yes |
| 115 | Lap3 | 12352 | -0.784 | -0.0194 | Yes |
| 116 | Usp16 | 12371 | -0.818 | 0.0057 | Yes |