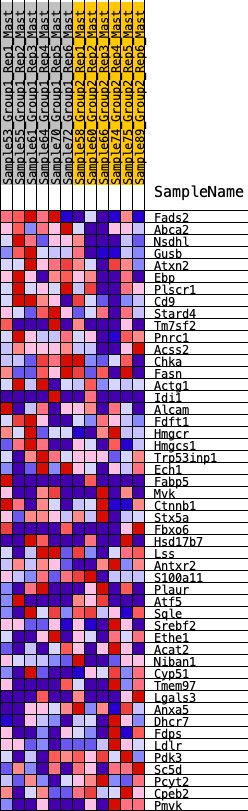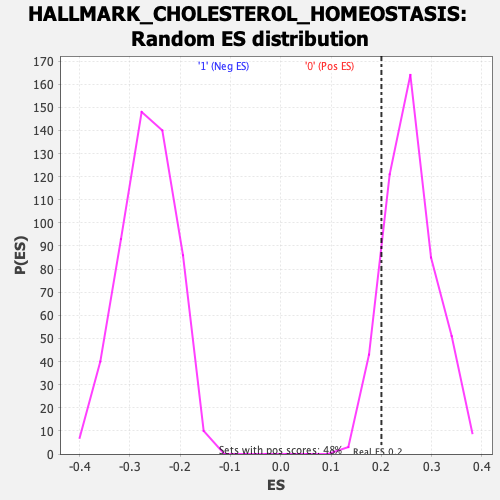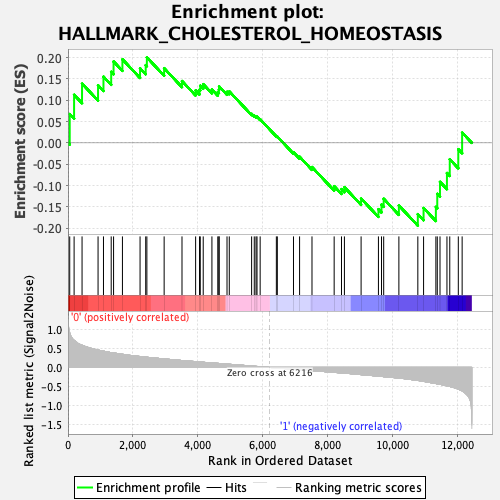
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.2002223 |
| Normalized Enrichment Score (NES) | 0.7748761 |
| Nominal p-value | 0.87815124 |
| FDR q-value | 0.9516799 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fads2 | 53 | 0.914 | 0.0675 | Yes |
| 2 | Abca2 | 188 | 0.713 | 0.1126 | Yes |
| 3 | Nsdhl | 433 | 0.585 | 0.1388 | Yes |
| 4 | Gusb | 926 | 0.456 | 0.1349 | Yes |
| 5 | Atxn2 | 1093 | 0.428 | 0.1550 | Yes |
| 6 | Ebp | 1331 | 0.390 | 0.1665 | Yes |
| 7 | Plscr1 | 1404 | 0.382 | 0.1906 | Yes |
| 8 | Cd9 | 1677 | 0.346 | 0.1958 | Yes |
| 9 | Stard4 | 2220 | 0.288 | 0.1747 | Yes |
| 10 | Tm7sf2 | 2394 | 0.271 | 0.1820 | Yes |
| 11 | Pnrc1 | 2430 | 0.268 | 0.2002 | Yes |
| 12 | Acss2 | 2962 | 0.223 | 0.1749 | No |
| 13 | Chka | 3512 | 0.182 | 0.1448 | No |
| 14 | Fasn | 3932 | 0.152 | 0.1229 | No |
| 15 | Actg1 | 4052 | 0.143 | 0.1246 | No |
| 16 | Idi1 | 4075 | 0.143 | 0.1340 | No |
| 17 | Alcam | 4166 | 0.136 | 0.1374 | No |
| 18 | Fdft1 | 4434 | 0.118 | 0.1251 | No |
| 19 | Hmgcr | 4615 | 0.107 | 0.1190 | No |
| 20 | Hmgcs1 | 4649 | 0.105 | 0.1245 | No |
| 21 | Trp53inp1 | 4656 | 0.104 | 0.1322 | No |
| 22 | Ech1 | 4899 | 0.088 | 0.1196 | No |
| 23 | Fabp5 | 4969 | 0.083 | 0.1205 | No |
| 24 | Mvk | 5657 | 0.036 | 0.0679 | No |
| 25 | Ctnnb1 | 5739 | 0.030 | 0.0638 | No |
| 26 | Stx5a | 5792 | 0.027 | 0.0617 | No |
| 27 | Fbxo6 | 5823 | 0.026 | 0.0613 | No |
| 28 | Hsd17b7 | 5920 | 0.019 | 0.0551 | No |
| 29 | Lss | 6419 | -0.012 | 0.0158 | No |
| 30 | Antxr2 | 6449 | -0.014 | 0.0146 | No |
| 31 | S100a11 | 6947 | -0.045 | -0.0220 | No |
| 32 | Plaur | 7135 | -0.057 | -0.0326 | No |
| 33 | Atf5 | 7517 | -0.084 | -0.0567 | No |
| 34 | Sqle | 8200 | -0.130 | -0.1016 | No |
| 35 | Srebf2 | 8426 | -0.144 | -0.1085 | No |
| 36 | Ethe1 | 8516 | -0.150 | -0.1039 | No |
| 37 | Acat2 | 9030 | -0.189 | -0.1305 | No |
| 38 | Niban1 | 9564 | -0.229 | -0.1555 | No |
| 39 | Cyp51 | 9659 | -0.236 | -0.1446 | No |
| 40 | Tmem97 | 9726 | -0.241 | -0.1310 | No |
| 41 | Lgals3 | 10194 | -0.279 | -0.1468 | No |
| 42 | Anxa5 | 10778 | -0.341 | -0.1671 | No |
| 43 | Dhcr7 | 10955 | -0.365 | -0.1527 | No |
| 44 | Fdps | 11334 | -0.423 | -0.1500 | No |
| 45 | Ldlr | 11377 | -0.431 | -0.1195 | No |
| 46 | Pdk3 | 11461 | -0.444 | -0.0914 | No |
| 47 | Sc5d | 11675 | -0.483 | -0.0707 | No |
| 48 | Pcyt2 | 11762 | -0.500 | -0.0384 | No |
| 49 | Cpeb2 | 12025 | -0.569 | -0.0149 | No |
| 50 | Pmvk | 12143 | -0.617 | 0.0240 | No |