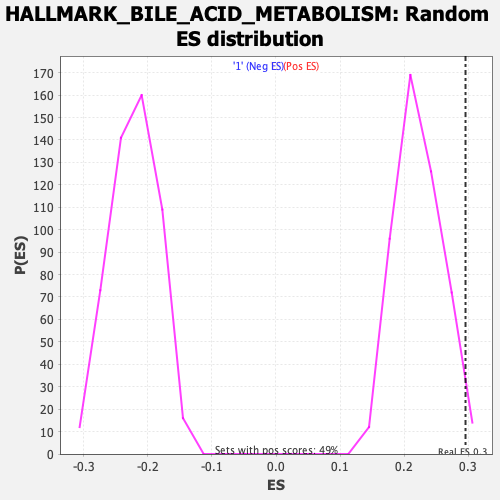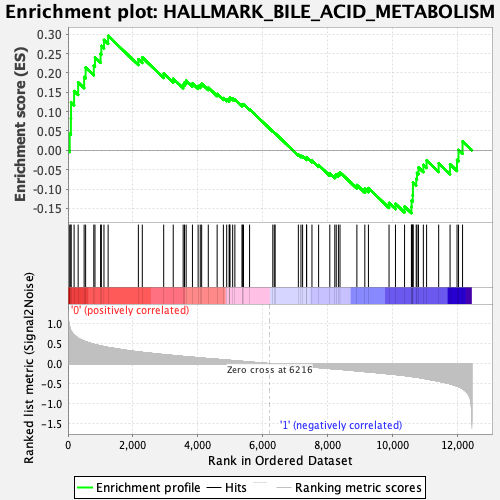
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.29558763 |
| Normalized Enrichment Score (NES) | 1.327992 |
| Nominal p-value | 0.010224949 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.666 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fads2 | 53 | 0.914 | 0.0427 | Yes |
| 2 | Gstk1 | 89 | 0.824 | 0.0822 | Yes |
| 3 | Pex19 | 94 | 0.820 | 0.1240 | Yes |
| 4 | Abca2 | 188 | 0.713 | 0.1531 | Yes |
| 5 | Aldh9a1 | 313 | 0.630 | 0.1754 | Yes |
| 6 | Soat2 | 497 | 0.561 | 0.1894 | Yes |
| 7 | Slc23a2 | 543 | 0.548 | 0.2139 | Yes |
| 8 | Pex7 | 795 | 0.482 | 0.2184 | Yes |
| 9 | Sod1 | 831 | 0.477 | 0.2401 | Yes |
| 10 | Hsd17b11 | 1004 | 0.443 | 0.2490 | Yes |
| 11 | Phyh | 1028 | 0.439 | 0.2697 | Yes |
| 12 | Pfkm | 1108 | 0.425 | 0.2851 | Yes |
| 13 | Cat | 1236 | 0.404 | 0.2956 | Yes |
| 14 | Aqp9 | 2168 | 0.292 | 0.2353 | No |
| 15 | Pex11g | 2289 | 0.281 | 0.2400 | No |
| 16 | Pnpla8 | 2948 | 0.224 | 0.1983 | No |
| 17 | Nr3c2 | 3242 | 0.202 | 0.1850 | No |
| 18 | Lipe | 3548 | 0.179 | 0.1695 | No |
| 19 | Nedd4 | 3589 | 0.176 | 0.1753 | No |
| 20 | Idh1 | 3640 | 0.172 | 0.1801 | No |
| 21 | Slc35b2 | 3834 | 0.159 | 0.1727 | No |
| 22 | Pxmp2 | 4009 | 0.147 | 0.1662 | No |
| 23 | Idi1 | 4075 | 0.143 | 0.1682 | No |
| 24 | Acsl1 | 4118 | 0.140 | 0.1720 | No |
| 25 | Pex13 | 4322 | 0.125 | 0.1620 | No |
| 26 | Hsd17b4 | 4595 | 0.107 | 0.1456 | No |
| 27 | Mlycd | 4787 | 0.095 | 0.1350 | No |
| 28 | Hacl1 | 4889 | 0.090 | 0.1315 | No |
| 29 | Pex11a | 4964 | 0.083 | 0.1297 | No |
| 30 | Isoc1 | 4980 | 0.082 | 0.1327 | No |
| 31 | Abca3 | 4989 | 0.081 | 0.1363 | No |
| 32 | Slc29a1 | 5073 | 0.075 | 0.1334 | No |
| 33 | Prdx5 | 5141 | 0.072 | 0.1317 | No |
| 34 | Aldh1a1 | 5364 | 0.056 | 0.1166 | No |
| 35 | Nudt12 | 5370 | 0.056 | 0.1191 | No |
| 36 | Abcd1 | 5407 | 0.054 | 0.1190 | No |
| 37 | Idh2 | 5594 | 0.041 | 0.1060 | No |
| 38 | Hsd3b7 | 6310 | -0.007 | 0.0486 | No |
| 39 | Paox | 6355 | -0.009 | 0.0455 | No |
| 40 | Pex16 | 6382 | -0.011 | 0.0439 | No |
| 41 | Acsl5 | 7097 | -0.055 | -0.0110 | No |
| 42 | Pex26 | 7177 | -0.060 | -0.0143 | No |
| 43 | Crot | 7228 | -0.063 | -0.0151 | No |
| 44 | Pex12 | 7349 | -0.072 | -0.0211 | No |
| 45 | Fads1 | 7353 | -0.072 | -0.0177 | No |
| 46 | Bcar3 | 7516 | -0.084 | -0.0264 | No |
| 47 | Abca5 | 7721 | -0.097 | -0.0379 | No |
| 48 | Dhcr24 | 8066 | -0.120 | -0.0596 | No |
| 49 | Npc1 | 8220 | -0.130 | -0.0652 | No |
| 50 | Pecr | 8264 | -0.133 | -0.0619 | No |
| 51 | Pex6 | 8331 | -0.137 | -0.0602 | No |
| 52 | Cyp39a1 | 8380 | -0.141 | -0.0568 | No |
| 53 | Pex1 | 8900 | -0.178 | -0.0896 | No |
| 54 | Slc22a18 | 9145 | -0.197 | -0.0992 | No |
| 55 | Klf1 | 9256 | -0.205 | -0.0976 | No |
| 56 | Optn | 9891 | -0.254 | -0.1358 | No |
| 57 | Abcd3 | 10089 | -0.270 | -0.1379 | No |
| 58 | Lonp2 | 10369 | -0.295 | -0.1453 | No |
| 59 | Atxn1 | 10580 | -0.319 | -0.1459 | No |
| 60 | Ar | 10587 | -0.320 | -0.1299 | No |
| 61 | Amacr | 10621 | -0.325 | -0.1159 | No |
| 62 | Retsat | 10627 | -0.325 | -0.0996 | No |
| 63 | Abca8b | 10628 | -0.325 | -0.0829 | No |
| 64 | Fdxr | 10728 | -0.335 | -0.0737 | No |
| 65 | Dio2 | 10752 | -0.338 | -0.0582 | No |
| 66 | Scp2 | 10798 | -0.344 | -0.0442 | No |
| 67 | Abcd2 | 10948 | -0.364 | -0.0375 | No |
| 68 | Gclm | 11048 | -0.380 | -0.0260 | No |
| 69 | Gnpat | 11420 | -0.437 | -0.0335 | No |
| 70 | Efhc1 | 11771 | -0.503 | -0.0360 | No |
| 71 | Rxra | 11986 | -0.559 | -0.0246 | No |
| 72 | Abca1 | 12032 | -0.570 | 0.0010 | No |
| 73 | Rbp1 | 12156 | -0.622 | 0.0230 | No |