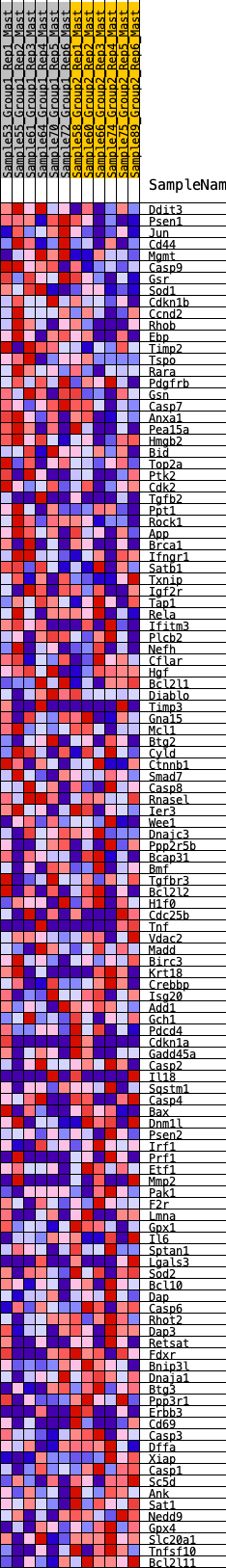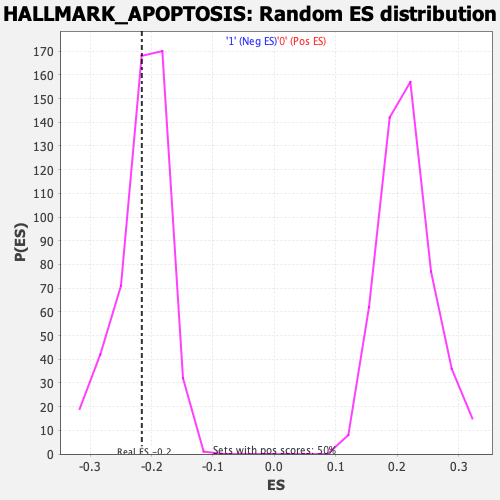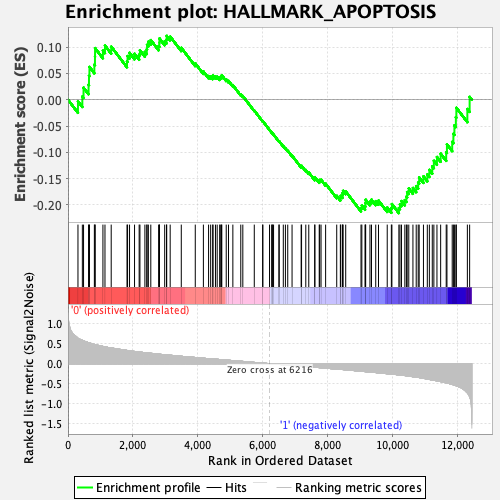
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.21578464 |
| Normalized Enrichment Score (NES) | -1.0008692 |
| Nominal p-value | 0.4413519 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ddit3 | 307 | 0.632 | -0.0030 | No |
| 2 | Psen1 | 442 | 0.581 | 0.0062 | No |
| 3 | Jun | 479 | 0.566 | 0.0229 | No |
| 4 | Cd44 | 634 | 0.522 | 0.0285 | No |
| 5 | Mgmt | 647 | 0.520 | 0.0456 | No |
| 6 | Casp9 | 661 | 0.517 | 0.0624 | No |
| 7 | Gsr | 812 | 0.480 | 0.0669 | No |
| 8 | Sod1 | 831 | 0.477 | 0.0819 | No |
| 9 | Cdkn1b | 834 | 0.476 | 0.0983 | No |
| 10 | Ccnd2 | 1076 | 0.430 | 0.0936 | No |
| 11 | Rhob | 1139 | 0.420 | 0.1031 | No |
| 12 | Ebp | 1331 | 0.390 | 0.1011 | No |
| 13 | Timp2 | 1818 | 0.330 | 0.0731 | No |
| 14 | Tspo | 1835 | 0.327 | 0.0831 | No |
| 15 | Rara | 1896 | 0.319 | 0.0893 | No |
| 16 | Pdgfrb | 2050 | 0.305 | 0.0874 | No |
| 17 | Gsn | 2192 | 0.290 | 0.0860 | No |
| 18 | Casp7 | 2216 | 0.288 | 0.0942 | No |
| 19 | Anxa1 | 2369 | 0.274 | 0.0913 | No |
| 20 | Pea15a | 2427 | 0.269 | 0.0960 | No |
| 21 | Hmgb2 | 2436 | 0.268 | 0.1046 | No |
| 22 | Bid | 2476 | 0.264 | 0.1106 | No |
| 23 | Top2a | 2551 | 0.257 | 0.1135 | No |
| 24 | Ptk2 | 2792 | 0.238 | 0.1023 | No |
| 25 | Cdk2 | 2816 | 0.235 | 0.1086 | No |
| 26 | Tgfb2 | 2818 | 0.235 | 0.1166 | No |
| 27 | Ppt1 | 2979 | 0.222 | 0.1113 | No |
| 28 | Rock1 | 3035 | 0.218 | 0.1144 | No |
| 29 | App | 3041 | 0.217 | 0.1215 | No |
| 30 | Brca1 | 3148 | 0.209 | 0.1202 | No |
| 31 | Ifngr1 | 3490 | 0.183 | 0.0988 | No |
| 32 | Satb1 | 3921 | 0.153 | 0.0692 | No |
| 33 | Txnip | 4171 | 0.136 | 0.0537 | No |
| 34 | Igf2r | 4331 | 0.124 | 0.0451 | No |
| 35 | Tap1 | 4395 | 0.121 | 0.0442 | No |
| 36 | Rela | 4459 | 0.116 | 0.0431 | No |
| 37 | Ifitm3 | 4467 | 0.115 | 0.0465 | No |
| 38 | Plcb2 | 4543 | 0.110 | 0.0443 | No |
| 39 | Nefh | 4596 | 0.107 | 0.0438 | No |
| 40 | Cflar | 4674 | 0.103 | 0.0411 | No |
| 41 | Hgf | 4691 | 0.102 | 0.0433 | No |
| 42 | Bcl2l1 | 4731 | 0.099 | 0.0436 | No |
| 43 | Diablo | 4737 | 0.098 | 0.0466 | No |
| 44 | Timp3 | 4875 | 0.090 | 0.0386 | No |
| 45 | Gna15 | 4945 | 0.084 | 0.0359 | No |
| 46 | Mcl1 | 5079 | 0.074 | 0.0277 | No |
| 47 | Btg2 | 5323 | 0.060 | 0.0101 | No |
| 48 | Cyld | 5386 | 0.056 | 0.0070 | No |
| 49 | Ctnnb1 | 5739 | 0.030 | -0.0205 | No |
| 50 | Smad7 | 5997 | 0.015 | -0.0409 | No |
| 51 | Casp8 | 6003 | 0.014 | -0.0408 | No |
| 52 | Rnasel | 6209 | 0.000 | -0.0574 | No |
| 53 | Ier3 | 6276 | -0.004 | -0.0627 | No |
| 54 | Wee1 | 6290 | -0.005 | -0.0635 | No |
| 55 | Dnajc3 | 6307 | -0.006 | -0.0646 | No |
| 56 | Ppp2r5b | 6333 | -0.008 | -0.0664 | No |
| 57 | Bcap31 | 6499 | -0.018 | -0.0791 | No |
| 58 | Bmf | 6506 | -0.018 | -0.0790 | No |
| 59 | Tgfbr3 | 6633 | -0.027 | -0.0883 | No |
| 60 | Bcl2l2 | 6698 | -0.030 | -0.0925 | No |
| 61 | H1f0 | 6770 | -0.034 | -0.0971 | No |
| 62 | Cdc25b | 6903 | -0.041 | -0.1064 | No |
| 63 | Tnf | 7183 | -0.061 | -0.1269 | No |
| 64 | Vdac2 | 7196 | -0.062 | -0.1257 | No |
| 65 | Madd | 7326 | -0.070 | -0.1338 | No |
| 66 | Birc3 | 7418 | -0.077 | -0.1385 | No |
| 67 | Krt18 | 7601 | -0.090 | -0.1502 | No |
| 68 | Crebbp | 7606 | -0.090 | -0.1474 | No |
| 69 | Isg20 | 7740 | -0.098 | -0.1548 | No |
| 70 | Add1 | 7754 | -0.099 | -0.1524 | No |
| 71 | Gch1 | 7796 | -0.100 | -0.1523 | No |
| 72 | Pdcd4 | 7937 | -0.111 | -0.1598 | No |
| 73 | Cdkn1a | 8279 | -0.134 | -0.1828 | No |
| 74 | Gadd45a | 8391 | -0.142 | -0.1869 | No |
| 75 | Casp2 | 8402 | -0.142 | -0.1828 | No |
| 76 | Il18 | 8455 | -0.146 | -0.1819 | No |
| 77 | Sqstm1 | 8461 | -0.146 | -0.1773 | No |
| 78 | Casp4 | 8475 | -0.147 | -0.1733 | No |
| 79 | Bax | 8556 | -0.152 | -0.1745 | No |
| 80 | Dnm1l | 9027 | -0.188 | -0.2061 | No |
| 81 | Psen2 | 9053 | -0.190 | -0.2015 | No |
| 82 | Irf1 | 9152 | -0.197 | -0.2026 | No |
| 83 | Prf1 | 9164 | -0.198 | -0.1967 | No |
| 84 | Etf1 | 9171 | -0.199 | -0.1903 | No |
| 85 | Mmp2 | 9303 | -0.208 | -0.1937 | No |
| 86 | Pak1 | 9350 | -0.212 | -0.1901 | No |
| 87 | F2r | 9481 | -0.221 | -0.1930 | No |
| 88 | Lmna | 9566 | -0.229 | -0.1919 | No |
| 89 | Gpx1 | 9835 | -0.250 | -0.2050 | No |
| 90 | Il6 | 9962 | -0.260 | -0.2062 | Yes |
| 91 | Sptan1 | 9979 | -0.261 | -0.1984 | Yes |
| 92 | Lgals3 | 10194 | -0.279 | -0.2061 | Yes |
| 93 | Sod2 | 10227 | -0.282 | -0.1990 | Yes |
| 94 | Bcl10 | 10274 | -0.287 | -0.1928 | Yes |
| 95 | Dap | 10378 | -0.296 | -0.1909 | Yes |
| 96 | Casp6 | 10426 | -0.301 | -0.1842 | Yes |
| 97 | Rhot2 | 10449 | -0.303 | -0.1755 | Yes |
| 98 | Dap3 | 10499 | -0.309 | -0.1688 | Yes |
| 99 | Retsat | 10627 | -0.325 | -0.1679 | Yes |
| 100 | Fdxr | 10728 | -0.335 | -0.1644 | Yes |
| 101 | Bnip3l | 10789 | -0.343 | -0.1574 | Yes |
| 102 | Dnaja1 | 10819 | -0.347 | -0.1477 | Yes |
| 103 | Btg3 | 10951 | -0.365 | -0.1457 | Yes |
| 104 | Ppp3r1 | 11070 | -0.383 | -0.1420 | Yes |
| 105 | Erbb3 | 11133 | -0.393 | -0.1334 | Yes |
| 106 | Cd69 | 11223 | -0.407 | -0.1266 | Yes |
| 107 | Casp3 | 11272 | -0.413 | -0.1162 | Yes |
| 108 | Dffa | 11367 | -0.429 | -0.1089 | Yes |
| 109 | Xiap | 11481 | -0.447 | -0.1026 | Yes |
| 110 | Casp1 | 11652 | -0.480 | -0.0998 | Yes |
| 111 | Sc5d | 11675 | -0.483 | -0.0848 | Yes |
| 112 | Ank | 11837 | -0.517 | -0.0800 | Yes |
| 113 | Sat1 | 11874 | -0.526 | -0.0647 | Yes |
| 114 | Nedd9 | 11905 | -0.534 | -0.0486 | Yes |
| 115 | Gpx4 | 11950 | -0.548 | -0.0332 | Yes |
| 116 | Slc20a1 | 11965 | -0.551 | -0.0153 | Yes |
| 117 | Tnfsf10 | 12304 | -0.726 | -0.0176 | Yes |
| 118 | Bcl2l11 | 12374 | -0.827 | 0.0054 | Yes |