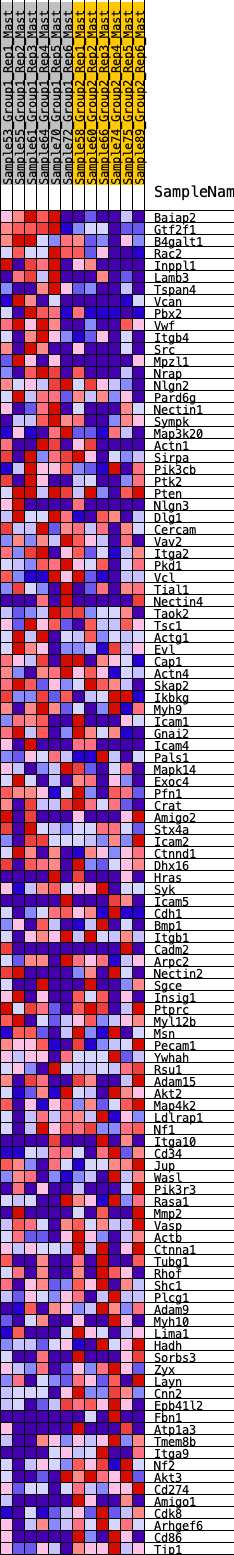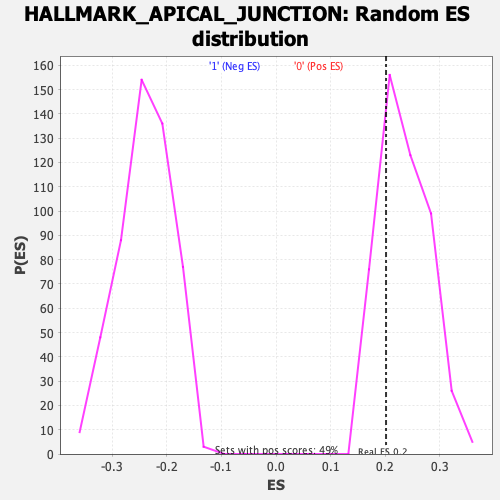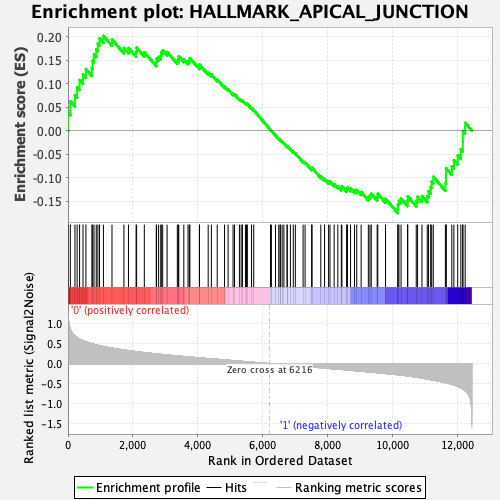
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | 0.20178223 |
| Normalized Enrichment Score (NES) | 0.85680693 |
| Nominal p-value | 0.7340206 |
| FDR q-value | 0.9418703 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Baiap2 | 11 | 1.111 | 0.0378 | Yes |
| 2 | Gtf2f1 | 72 | 0.849 | 0.0624 | Yes |
| 3 | B4galt1 | 214 | 0.698 | 0.0753 | Yes |
| 4 | Rac2 | 281 | 0.652 | 0.0926 | Yes |
| 5 | Inppl1 | 357 | 0.611 | 0.1078 | Yes |
| 6 | Lamb3 | 462 | 0.573 | 0.1193 | Yes |
| 7 | Tspan4 | 552 | 0.547 | 0.1311 | Yes |
| 8 | Vcan | 737 | 0.497 | 0.1335 | Yes |
| 9 | Pbx2 | 760 | 0.490 | 0.1487 | Yes |
| 10 | Vwf | 801 | 0.482 | 0.1623 | Yes |
| 11 | Itgb4 | 870 | 0.468 | 0.1730 | Yes |
| 12 | Src | 920 | 0.457 | 0.1850 | Yes |
| 13 | Mpzl1 | 972 | 0.449 | 0.1965 | Yes |
| 14 | Nrap | 1091 | 0.428 | 0.2018 | Yes |
| 15 | Nlgn2 | 1355 | 0.388 | 0.1939 | No |
| 16 | Pard6g | 1723 | 0.341 | 0.1761 | No |
| 17 | Nectin1 | 1863 | 0.323 | 0.1760 | No |
| 18 | Sympk | 2098 | 0.299 | 0.1675 | No |
| 19 | Map3k20 | 2114 | 0.298 | 0.1766 | No |
| 20 | Actn1 | 2351 | 0.275 | 0.1671 | No |
| 21 | Sirpa | 2719 | 0.244 | 0.1458 | No |
| 22 | Pik3cb | 2728 | 0.243 | 0.1536 | No |
| 23 | Ptk2 | 2792 | 0.238 | 0.1568 | No |
| 24 | Pten | 2860 | 0.231 | 0.1594 | No |
| 25 | Nlgn3 | 2872 | 0.230 | 0.1665 | No |
| 26 | Dlg1 | 2917 | 0.226 | 0.1708 | No |
| 27 | Cercam | 3053 | 0.217 | 0.1674 | No |
| 28 | Vav2 | 3368 | 0.192 | 0.1486 | No |
| 29 | Itga2 | 3401 | 0.190 | 0.1526 | No |
| 30 | Pkd1 | 3413 | 0.189 | 0.1583 | No |
| 31 | Vcl | 3568 | 0.177 | 0.1520 | No |
| 32 | Tial1 | 3698 | 0.169 | 0.1474 | No |
| 33 | Nectin4 | 3737 | 0.167 | 0.1501 | No |
| 34 | Taok2 | 3755 | 0.165 | 0.1545 | No |
| 35 | Tsc1 | 4047 | 0.144 | 0.1359 | No |
| 36 | Actg1 | 4052 | 0.143 | 0.1406 | No |
| 37 | Evl | 4319 | 0.125 | 0.1234 | No |
| 38 | Cap1 | 4417 | 0.119 | 0.1196 | No |
| 39 | Actn4 | 4597 | 0.107 | 0.1088 | No |
| 40 | Skap2 | 4823 | 0.093 | 0.0938 | No |
| 41 | Ikbkg | 4932 | 0.086 | 0.0881 | No |
| 42 | Myh9 | 5085 | 0.074 | 0.0783 | No |
| 43 | Icam1 | 5130 | 0.072 | 0.0773 | No |
| 44 | Gnai2 | 5281 | 0.062 | 0.0672 | No |
| 45 | Icam4 | 5334 | 0.059 | 0.0651 | No |
| 46 | Pals1 | 5374 | 0.056 | 0.0639 | No |
| 47 | Mapk14 | 5467 | 0.049 | 0.0581 | No |
| 48 | Exoc4 | 5499 | 0.047 | 0.0572 | No |
| 49 | Pfn1 | 5526 | 0.045 | 0.0567 | No |
| 50 | Crat | 5655 | 0.036 | 0.0476 | No |
| 51 | Amigo2 | 5722 | 0.032 | 0.0433 | No |
| 52 | Stx4a | 6238 | -0.001 | 0.0016 | No |
| 53 | Icam2 | 6266 | -0.003 | -0.0005 | No |
| 54 | Ctnnd1 | 6393 | -0.011 | -0.0103 | No |
| 55 | Dhx16 | 6493 | -0.017 | -0.0177 | No |
| 56 | Hras | 6537 | -0.021 | -0.0205 | No |
| 57 | Syk | 6540 | -0.021 | -0.0199 | No |
| 58 | Icam5 | 6598 | -0.024 | -0.0237 | No |
| 59 | Cdh1 | 6651 | -0.027 | -0.0270 | No |
| 60 | Bmp1 | 6751 | -0.033 | -0.0339 | No |
| 61 | Itgb1 | 6753 | -0.033 | -0.0328 | No |
| 62 | Cadm2 | 6852 | -0.038 | -0.0394 | No |
| 63 | Arpc2 | 6940 | -0.044 | -0.0450 | No |
| 64 | Nectin2 | 7004 | -0.048 | -0.0484 | No |
| 65 | Sgce | 7243 | -0.065 | -0.0654 | No |
| 66 | Insig1 | 7304 | -0.069 | -0.0679 | No |
| 67 | Ptprc | 7506 | -0.083 | -0.0813 | No |
| 68 | Myl12b | 7519 | -0.085 | -0.0793 | No |
| 69 | Msn | 7788 | -0.100 | -0.0976 | No |
| 70 | Pecam1 | 7904 | -0.108 | -0.1031 | No |
| 71 | Ywhah | 8029 | -0.118 | -0.1091 | No |
| 72 | Rsu1 | 8072 | -0.121 | -0.1083 | No |
| 73 | Adam15 | 8199 | -0.129 | -0.1140 | No |
| 74 | Akt2 | 8313 | -0.136 | -0.1184 | No |
| 75 | Map4k2 | 8420 | -0.143 | -0.1220 | No |
| 76 | Ldlrap1 | 8438 | -0.145 | -0.1184 | No |
| 77 | Nf1 | 8583 | -0.155 | -0.1247 | No |
| 78 | Itga10 | 8612 | -0.156 | -0.1215 | No |
| 79 | Cd34 | 8705 | -0.162 | -0.1233 | No |
| 80 | Jup | 8827 | -0.173 | -0.1271 | No |
| 81 | Wasl | 8896 | -0.178 | -0.1264 | No |
| 82 | Pik3r3 | 9031 | -0.189 | -0.1307 | No |
| 83 | Rasa1 | 9253 | -0.204 | -0.1415 | No |
| 84 | Mmp2 | 9303 | -0.208 | -0.1383 | No |
| 85 | Vasp | 9343 | -0.211 | -0.1341 | No |
| 86 | Actb | 9525 | -0.225 | -0.1409 | No |
| 87 | Ctnna1 | 9545 | -0.227 | -0.1346 | No |
| 88 | Tubg1 | 9783 | -0.246 | -0.1452 | No |
| 89 | Rhof | 10161 | -0.277 | -0.1662 | No |
| 90 | Shc1 | 10165 | -0.277 | -0.1568 | No |
| 91 | Plcg1 | 10196 | -0.279 | -0.1495 | No |
| 92 | Adam9 | 10261 | -0.285 | -0.1448 | No |
| 93 | Myh10 | 10462 | -0.304 | -0.1504 | No |
| 94 | Lima1 | 10467 | -0.305 | -0.1401 | No |
| 95 | Hadh | 10729 | -0.335 | -0.1496 | No |
| 96 | Sorbs3 | 10770 | -0.340 | -0.1410 | No |
| 97 | Zyx | 10905 | -0.360 | -0.1394 | No |
| 98 | Layn | 11068 | -0.383 | -0.1392 | No |
| 99 | Cnn2 | 11108 | -0.389 | -0.1289 | No |
| 100 | Epb41l2 | 11174 | -0.400 | -0.1202 | No |
| 101 | Fbn1 | 11205 | -0.404 | -0.1086 | No |
| 102 | Atp1a3 | 11254 | -0.411 | -0.0982 | No |
| 103 | Tmem8b | 11627 | -0.476 | -0.1118 | No |
| 104 | Itga9 | 11648 | -0.479 | -0.0968 | No |
| 105 | Nf2 | 11649 | -0.480 | -0.0801 | No |
| 106 | Akt3 | 11825 | -0.515 | -0.0763 | No |
| 107 | Cd274 | 11889 | -0.530 | -0.0630 | No |
| 108 | Amigo1 | 12006 | -0.565 | -0.0528 | No |
| 109 | Cdk8 | 12103 | -0.598 | -0.0398 | No |
| 110 | Arhgef6 | 12163 | -0.625 | -0.0228 | No |
| 111 | Cd86 | 12165 | -0.627 | -0.0011 | No |
| 112 | Tjp1 | 12236 | -0.672 | 0.0166 | No |