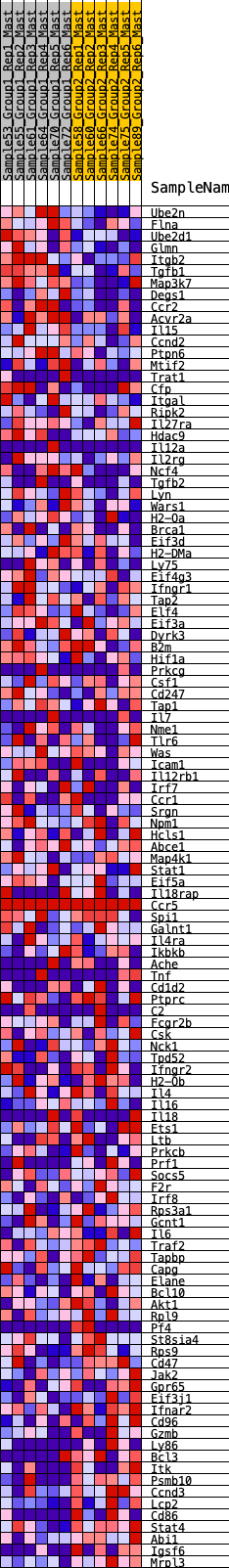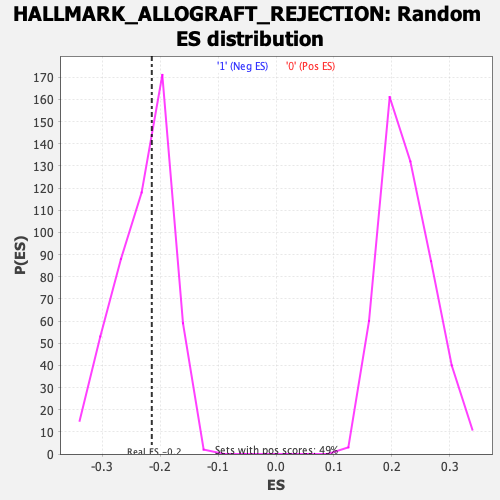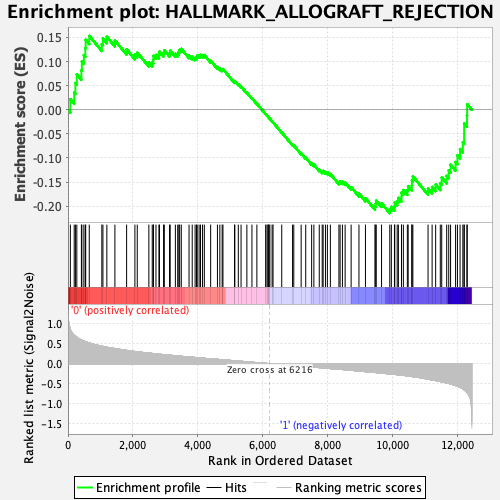
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group1_versus_Group2.Mast_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Mast_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_ALLOGRAFT_REJECTION |
| Enrichment Score (ES) | -0.21445522 |
| Normalized Enrichment Score (NES) | -0.94636464 |
| Nominal p-value | 0.5395257 |
| FDR q-value | 0.82095987 |
| FWER p-Value | 0.994 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ube2n | 73 | 0.849 | 0.0219 | No |
| 2 | Flna | 191 | 0.713 | 0.0358 | No |
| 3 | Ube2d1 | 229 | 0.688 | 0.0554 | No |
| 4 | Glmn | 272 | 0.659 | 0.0736 | No |
| 5 | Itgb2 | 413 | 0.593 | 0.0818 | No |
| 6 | Tgfb1 | 427 | 0.586 | 0.0999 | No |
| 7 | Map3k7 | 489 | 0.563 | 0.1135 | No |
| 8 | Degs1 | 534 | 0.551 | 0.1280 | No |
| 9 | Ccr2 | 545 | 0.548 | 0.1452 | No |
| 10 | Acvr2a | 655 | 0.518 | 0.1533 | No |
| 11 | Il15 | 1042 | 0.436 | 0.1363 | No |
| 12 | Ccnd2 | 1076 | 0.430 | 0.1478 | No |
| 13 | Ptpn6 | 1197 | 0.410 | 0.1515 | No |
| 14 | Mtif2 | 1445 | 0.377 | 0.1438 | No |
| 15 | Trat1 | 1806 | 0.330 | 0.1255 | No |
| 16 | Cfp | 2061 | 0.303 | 0.1148 | No |
| 17 | Itgal | 2134 | 0.295 | 0.1186 | No |
| 18 | Ripk2 | 2491 | 0.263 | 0.0984 | No |
| 19 | Il27ra | 2595 | 0.255 | 0.0984 | No |
| 20 | Hdac9 | 2629 | 0.252 | 0.1040 | No |
| 21 | Il12a | 2632 | 0.251 | 0.1120 | No |
| 22 | Il2rg | 2710 | 0.245 | 0.1138 | No |
| 23 | Ncf4 | 2802 | 0.237 | 0.1142 | No |
| 24 | Tgfb2 | 2818 | 0.235 | 0.1207 | No |
| 25 | Lyn | 2944 | 0.224 | 0.1179 | No |
| 26 | Wars1 | 2967 | 0.222 | 0.1235 | No |
| 27 | H2-Oa | 3128 | 0.211 | 0.1174 | No |
| 28 | Brca1 | 3148 | 0.209 | 0.1227 | No |
| 29 | Eif3d | 3308 | 0.197 | 0.1163 | No |
| 30 | H2-DMa | 3380 | 0.191 | 0.1168 | No |
| 31 | Ly75 | 3415 | 0.189 | 0.1202 | No |
| 32 | Eif4g3 | 3440 | 0.188 | 0.1245 | No |
| 33 | Ifngr1 | 3490 | 0.183 | 0.1265 | No |
| 34 | Tap2 | 3729 | 0.167 | 0.1127 | No |
| 35 | Elf4 | 3827 | 0.160 | 0.1100 | No |
| 36 | Eif3a | 3916 | 0.153 | 0.1079 | No |
| 37 | Dyrk3 | 3959 | 0.150 | 0.1094 | No |
| 38 | B2m | 3981 | 0.148 | 0.1126 | No |
| 39 | Hif1a | 4039 | 0.144 | 0.1127 | No |
| 40 | Prkcg | 4083 | 0.143 | 0.1139 | No |
| 41 | Csf1 | 4145 | 0.138 | 0.1135 | No |
| 42 | Cd247 | 4200 | 0.133 | 0.1135 | No |
| 43 | Tap1 | 4395 | 0.121 | 0.1017 | No |
| 44 | Il7 | 4604 | 0.107 | 0.0883 | No |
| 45 | Nme1 | 4678 | 0.103 | 0.0858 | No |
| 46 | Tlr6 | 4749 | 0.098 | 0.0833 | No |
| 47 | Was | 4774 | 0.096 | 0.0845 | No |
| 48 | Icam1 | 5130 | 0.072 | 0.0581 | No |
| 49 | Il12rb1 | 5140 | 0.072 | 0.0597 | No |
| 50 | Irf7 | 5247 | 0.065 | 0.0532 | No |
| 51 | Ccr1 | 5332 | 0.059 | 0.0484 | No |
| 52 | Srgn | 5511 | 0.046 | 0.0355 | No |
| 53 | Npm1 | 5667 | 0.035 | 0.0240 | No |
| 54 | Hcls1 | 5819 | 0.026 | 0.0126 | No |
| 55 | Abce1 | 6089 | 0.008 | -0.0089 | No |
| 56 | Map4k1 | 6131 | 0.005 | -0.0121 | No |
| 57 | Stat1 | 6159 | 0.003 | -0.0142 | No |
| 58 | Eif5a | 6182 | 0.002 | -0.0159 | No |
| 59 | Il18rap | 6192 | 0.002 | -0.0166 | No |
| 60 | Ccr5 | 6219 | 0.000 | -0.0187 | No |
| 61 | Spi1 | 6288 | -0.005 | -0.0240 | No |
| 62 | Galnt1 | 6317 | -0.007 | -0.0261 | No |
| 63 | Il4ra | 6585 | -0.024 | -0.0469 | No |
| 64 | Ikbkb | 6918 | -0.042 | -0.0725 | No |
| 65 | Ache | 6956 | -0.046 | -0.0740 | No |
| 66 | Tnf | 7183 | -0.061 | -0.0903 | No |
| 67 | Cd1d2 | 7324 | -0.070 | -0.0994 | No |
| 68 | Ptprc | 7506 | -0.083 | -0.1113 | No |
| 69 | C2 | 7575 | -0.088 | -0.1140 | No |
| 70 | Fcgr2b | 7741 | -0.098 | -0.1241 | No |
| 71 | Csk | 7832 | -0.104 | -0.1280 | No |
| 72 | Nck1 | 7860 | -0.105 | -0.1268 | No |
| 73 | Tpd52 | 7932 | -0.111 | -0.1289 | No |
| 74 | Ifngr2 | 7991 | -0.115 | -0.1298 | No |
| 75 | H2-Ob | 8083 | -0.122 | -0.1332 | No |
| 76 | Il4 | 8347 | -0.138 | -0.1500 | No |
| 77 | Il16 | 8383 | -0.141 | -0.1482 | No |
| 78 | Il18 | 8455 | -0.146 | -0.1492 | No |
| 79 | Ets1 | 8538 | -0.151 | -0.1509 | No |
| 80 | Ltb | 8723 | -0.164 | -0.1604 | No |
| 81 | Prkcb | 8964 | -0.183 | -0.1739 | No |
| 82 | Prf1 | 9164 | -0.198 | -0.1835 | No |
| 83 | Socs5 | 9456 | -0.219 | -0.1999 | No |
| 84 | F2r | 9481 | -0.221 | -0.1946 | No |
| 85 | Irf8 | 9495 | -0.223 | -0.1884 | No |
| 86 | Rps3a1 | 9660 | -0.236 | -0.1939 | No |
| 87 | Gcnt1 | 9914 | -0.255 | -0.2061 | Yes |
| 88 | Il6 | 9962 | -0.260 | -0.2013 | Yes |
| 89 | Traf2 | 10058 | -0.268 | -0.2003 | Yes |
| 90 | Tapbp | 10068 | -0.268 | -0.1922 | Yes |
| 91 | Capg | 10142 | -0.275 | -0.1891 | Yes |
| 92 | Elane | 10179 | -0.278 | -0.1829 | Yes |
| 93 | Bcl10 | 10274 | -0.287 | -0.1811 | Yes |
| 94 | Akt1 | 10275 | -0.287 | -0.1717 | Yes |
| 95 | Rpl9 | 10332 | -0.292 | -0.1666 | Yes |
| 96 | Pf4 | 10455 | -0.304 | -0.1666 | Yes |
| 97 | St8sia4 | 10480 | -0.307 | -0.1584 | Yes |
| 98 | Rps9 | 10593 | -0.321 | -0.1570 | Yes |
| 99 | Cd47 | 10596 | -0.321 | -0.1466 | Yes |
| 100 | Jak2 | 10625 | -0.325 | -0.1382 | Yes |
| 101 | Gpr65 | 11093 | -0.386 | -0.1634 | Yes |
| 102 | Eif3j1 | 11218 | -0.406 | -0.1602 | Yes |
| 103 | Ifnar2 | 11327 | -0.422 | -0.1551 | Yes |
| 104 | Cd96 | 11470 | -0.446 | -0.1520 | Yes |
| 105 | Gzmb | 11514 | -0.453 | -0.1406 | Yes |
| 106 | Ly86 | 11667 | -0.482 | -0.1371 | Yes |
| 107 | Bcl3 | 11730 | -0.493 | -0.1259 | Yes |
| 108 | Itk | 11787 | -0.506 | -0.1139 | Yes |
| 109 | Psmb10 | 11941 | -0.546 | -0.1084 | Yes |
| 110 | Ccnd3 | 11998 | -0.562 | -0.0944 | Yes |
| 111 | Lcp2 | 12079 | -0.589 | -0.0816 | Yes |
| 112 | Cd86 | 12165 | -0.627 | -0.0679 | Yes |
| 113 | Stat4 | 12205 | -0.650 | -0.0497 | Yes |
| 114 | Abi1 | 12206 | -0.651 | -0.0284 | Yes |
| 115 | Igsf6 | 12292 | -0.718 | -0.0117 | Yes |
| 116 | Mrpl3 | 12298 | -0.723 | 0.0116 | Yes |