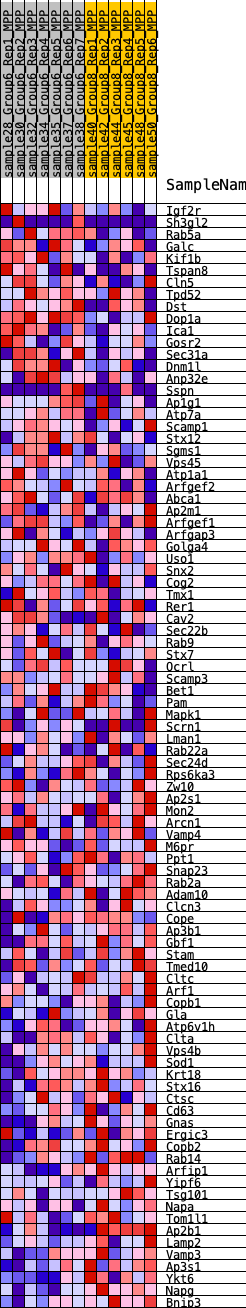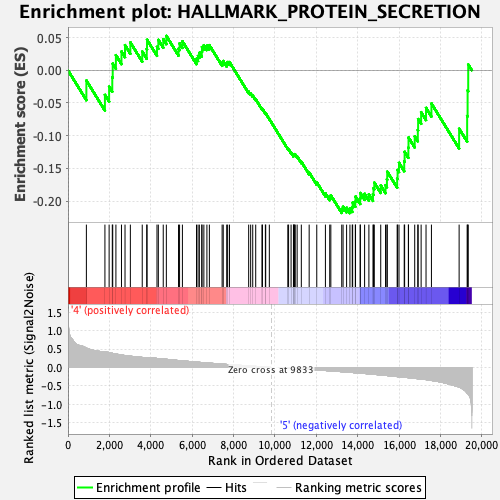
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group6_versus_Group8.MPP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | MPP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.21845053 |
| Normalized Enrichment Score (NES) | -1.1329117 |
| Nominal p-value | 0.27235773 |
| FDR q-value | 0.5183198 |
| FWER p-Value | 0.969 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Igf2r | 888 | 0.526 | -0.0156 | No |
| 2 | Sh3gl2 | 1784 | 0.420 | -0.0376 | No |
| 3 | Rab5a | 1987 | 0.403 | -0.0248 | No |
| 4 | Galc | 2140 | 0.384 | -0.0106 | No |
| 5 | Kif1b | 2161 | 0.381 | 0.0102 | No |
| 6 | Tspan8 | 2315 | 0.364 | 0.0232 | No |
| 7 | Cln5 | 2584 | 0.338 | 0.0288 | No |
| 8 | Tpd52 | 2755 | 0.323 | 0.0385 | No |
| 9 | Dst | 3013 | 0.303 | 0.0427 | No |
| 10 | Dop1a | 3587 | 0.272 | 0.0288 | No |
| 11 | Ica1 | 3805 | 0.261 | 0.0326 | No |
| 12 | Gosr2 | 3819 | 0.260 | 0.0468 | No |
| 13 | Sec31a | 4300 | 0.244 | 0.0361 | No |
| 14 | Dnm1l | 4365 | 0.240 | 0.0466 | No |
| 15 | Anp32e | 4602 | 0.231 | 0.0476 | No |
| 16 | Sspn | 4752 | 0.221 | 0.0527 | No |
| 17 | Ap1g1 | 5341 | 0.187 | 0.0331 | No |
| 18 | Atp7a | 5391 | 0.185 | 0.0412 | No |
| 19 | Scamp1 | 5528 | 0.178 | 0.0444 | No |
| 20 | Stx12 | 6209 | 0.146 | 0.0177 | No |
| 21 | Sgms1 | 6283 | 0.142 | 0.0221 | No |
| 22 | Vps45 | 6347 | 0.139 | 0.0268 | No |
| 23 | Atp1a1 | 6464 | 0.133 | 0.0285 | No |
| 24 | Arfgef2 | 6474 | 0.133 | 0.0356 | No |
| 25 | Abca1 | 6565 | 0.129 | 0.0384 | No |
| 26 | Ap2m1 | 6712 | 0.121 | 0.0378 | No |
| 27 | Arfgef1 | 6831 | 0.117 | 0.0385 | No |
| 28 | Arfgap3 | 7447 | 0.090 | 0.0120 | No |
| 29 | Golga4 | 7501 | 0.088 | 0.0143 | No |
| 30 | Uso1 | 7669 | 0.082 | 0.0104 | No |
| 31 | Snx2 | 7714 | 0.080 | 0.0127 | No |
| 32 | Cog2 | 7802 | 0.077 | 0.0126 | No |
| 33 | Tmx1 | 8730 | 0.042 | -0.0327 | No |
| 34 | Rer1 | 8826 | 0.038 | -0.0355 | No |
| 35 | Cav2 | 8926 | 0.034 | -0.0386 | No |
| 36 | Sec22b | 9070 | 0.029 | -0.0443 | No |
| 37 | Rab9 | 9372 | 0.017 | -0.0588 | No |
| 38 | Stx7 | 9397 | 0.016 | -0.0591 | No |
| 39 | Ocrl | 9537 | 0.011 | -0.0656 | No |
| 40 | Scamp3 | 9553 | 0.010 | -0.0658 | No |
| 41 | Bet1 | 9728 | 0.004 | -0.0746 | No |
| 42 | Pam | 10622 | -0.014 | -0.1198 | No |
| 43 | Mapk1 | 10658 | -0.014 | -0.1208 | No |
| 44 | Scrn1 | 10779 | -0.019 | -0.1259 | No |
| 45 | Lman1 | 10894 | -0.024 | -0.1304 | No |
| 46 | Rab22a | 10911 | -0.025 | -0.1298 | No |
| 47 | Sec24d | 10922 | -0.025 | -0.1289 | No |
| 48 | Rps6ka3 | 10939 | -0.026 | -0.1282 | No |
| 49 | Zw10 | 10980 | -0.027 | -0.1287 | No |
| 50 | Ap2s1 | 11072 | -0.031 | -0.1316 | No |
| 51 | Mon2 | 11274 | -0.038 | -0.1398 | No |
| 52 | Arcn1 | 11653 | -0.052 | -0.1563 | No |
| 53 | Vamp4 | 12020 | -0.067 | -0.1713 | No |
| 54 | M6pr | 12438 | -0.081 | -0.1881 | No |
| 55 | Ppt1 | 12647 | -0.089 | -0.1937 | No |
| 56 | Snap23 | 12700 | -0.091 | -0.1911 | No |
| 57 | Rab2a | 13227 | -0.112 | -0.2118 | Yes |
| 58 | Adam10 | 13291 | -0.115 | -0.2084 | Yes |
| 59 | Clcn3 | 13462 | -0.122 | -0.2102 | Yes |
| 60 | Cope | 13623 | -0.129 | -0.2110 | Yes |
| 61 | Ap3b1 | 13735 | -0.135 | -0.2090 | Yes |
| 62 | Gbf1 | 13755 | -0.136 | -0.2022 | Yes |
| 63 | Stam | 13883 | -0.141 | -0.2007 | Yes |
| 64 | Tmed10 | 13893 | -0.141 | -0.1931 | Yes |
| 65 | Cltc | 14118 | -0.151 | -0.1959 | Yes |
| 66 | Arf1 | 14126 | -0.152 | -0.1876 | Yes |
| 67 | Copb1 | 14332 | -0.162 | -0.1889 | Yes |
| 68 | Gla | 14539 | -0.171 | -0.1897 | Yes |
| 69 | Atp6v1h | 14737 | -0.180 | -0.1895 | Yes |
| 70 | Clta | 14762 | -0.181 | -0.1803 | Yes |
| 71 | Vps4b | 14800 | -0.184 | -0.1717 | Yes |
| 72 | Sod1 | 15111 | -0.202 | -0.1761 | Yes |
| 73 | Krt18 | 15343 | -0.214 | -0.1757 | Yes |
| 74 | Stx16 | 15411 | -0.217 | -0.1667 | Yes |
| 75 | Ctsc | 15425 | -0.218 | -0.1549 | Yes |
| 76 | Cd63 | 15905 | -0.246 | -0.1655 | Yes |
| 77 | Gnas | 15926 | -0.247 | -0.1524 | Yes |
| 78 | Ergic3 | 15994 | -0.250 | -0.1415 | Yes |
| 79 | Copb2 | 16246 | -0.267 | -0.1391 | Yes |
| 80 | Rab14 | 16266 | -0.269 | -0.1246 | Yes |
| 81 | Arfip1 | 16446 | -0.275 | -0.1181 | Yes |
| 82 | Yipf6 | 16454 | -0.275 | -0.1027 | Yes |
| 83 | Tsg101 | 16753 | -0.296 | -0.1011 | Yes |
| 84 | Napa | 16903 | -0.304 | -0.0913 | Yes |
| 85 | Tom1l1 | 16918 | -0.306 | -0.0745 | Yes |
| 86 | Ap2b1 | 17063 | -0.312 | -0.0640 | Yes |
| 87 | Lamp2 | 17300 | -0.327 | -0.0574 | Yes |
| 88 | Vamp3 | 17561 | -0.351 | -0.0507 | Yes |
| 89 | Ap3s1 | 18899 | -0.529 | -0.0892 | Yes |
| 90 | Ykt6 | 19287 | -0.688 | -0.0697 | Yes |
| 91 | Napg | 19309 | -0.698 | -0.0307 | Yes |
| 92 | Bnip3 | 19339 | -0.717 | 0.0089 | Yes |