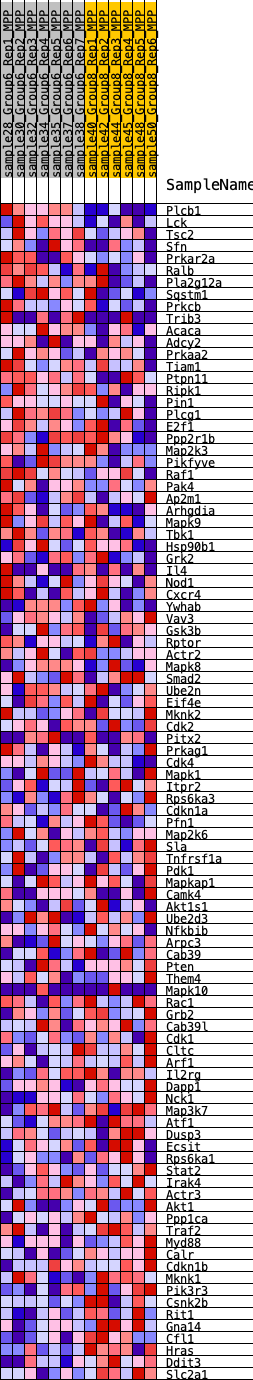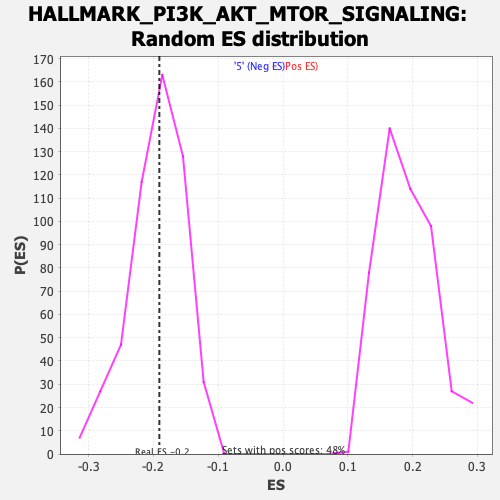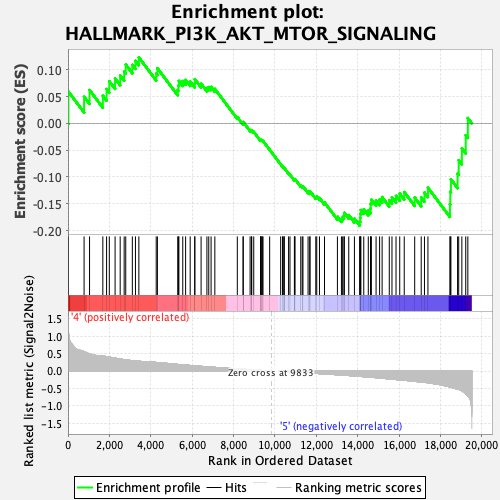
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group6_versus_Group8.MPP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | MPP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.19087327 |
| Normalized Enrichment Score (NES) | -0.9840485 |
| Nominal p-value | 0.48653847 |
| FDR q-value | 0.7293737 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Plcb1 | 21 | 1.139 | 0.0591 | No |
| 2 | Lck | 777 | 0.558 | 0.0497 | No |
| 3 | Tsc2 | 1041 | 0.493 | 0.0622 | No |
| 4 | Sfn | 1683 | 0.423 | 0.0515 | No |
| 5 | Prkar2a | 1864 | 0.412 | 0.0640 | No |
| 6 | Ralb | 1996 | 0.402 | 0.0785 | No |
| 7 | Pla2g12a | 2275 | 0.369 | 0.0837 | No |
| 8 | Sqstm1 | 2521 | 0.345 | 0.0893 | No |
| 9 | Prkcb | 2714 | 0.326 | 0.0966 | No |
| 10 | Trib3 | 2790 | 0.320 | 0.1097 | No |
| 11 | Acaca | 3109 | 0.299 | 0.1091 | No |
| 12 | Adcy2 | 3259 | 0.290 | 0.1167 | No |
| 13 | Prkaa2 | 3424 | 0.279 | 0.1230 | No |
| 14 | Tiam1 | 4262 | 0.246 | 0.0929 | No |
| 15 | Ptpn11 | 4320 | 0.243 | 0.1028 | No |
| 16 | Ripk1 | 5303 | 0.189 | 0.0622 | No |
| 17 | Pin1 | 5335 | 0.188 | 0.0705 | No |
| 18 | Plcg1 | 5358 | 0.186 | 0.0792 | No |
| 19 | E2f1 | 5545 | 0.177 | 0.0790 | No |
| 20 | Ppp2r1b | 5683 | 0.171 | 0.0810 | No |
| 21 | Map2k3 | 5904 | 0.160 | 0.0781 | No |
| 22 | Pikfyve | 6124 | 0.150 | 0.0747 | No |
| 23 | Raf1 | 6129 | 0.149 | 0.0824 | No |
| 24 | Pak4 | 6431 | 0.135 | 0.0740 | No |
| 25 | Ap2m1 | 6712 | 0.121 | 0.0660 | No |
| 26 | Arhgdia | 6801 | 0.118 | 0.0677 | No |
| 27 | Mapk9 | 6914 | 0.113 | 0.0680 | No |
| 28 | Tbk1 | 7095 | 0.105 | 0.0643 | No |
| 29 | Hsp90b1 | 8181 | 0.062 | 0.0116 | No |
| 30 | Grk2 | 8461 | 0.052 | 0.0000 | No |
| 31 | Il4 | 8466 | 0.052 | 0.0025 | No |
| 32 | Nod1 | 8799 | 0.039 | -0.0125 | No |
| 33 | Cxcr4 | 8865 | 0.036 | -0.0139 | No |
| 34 | Ywhab | 8879 | 0.036 | -0.0127 | No |
| 35 | Vav3 | 8975 | 0.032 | -0.0159 | No |
| 36 | Gsk3b | 9303 | 0.020 | -0.0317 | No |
| 37 | Rptor | 9314 | 0.020 | -0.0312 | No |
| 38 | Actr2 | 9327 | 0.019 | -0.0308 | No |
| 39 | Mapk8 | 9362 | 0.018 | -0.0316 | No |
| 40 | Smad2 | 9390 | 0.017 | -0.0321 | No |
| 41 | Ube2n | 9423 | 0.015 | -0.0330 | No |
| 42 | Eif4e | 9744 | 0.003 | -0.0493 | No |
| 43 | Mknk2 | 10276 | -0.004 | -0.0764 | No |
| 44 | Cdk2 | 10366 | -0.007 | -0.0807 | No |
| 45 | Pitx2 | 10394 | -0.007 | -0.0817 | No |
| 46 | Prkag1 | 10442 | -0.009 | -0.0836 | No |
| 47 | Cdk4 | 10448 | -0.009 | -0.0834 | No |
| 48 | Mapk1 | 10658 | -0.014 | -0.0934 | No |
| 49 | Itpr2 | 10726 | -0.017 | -0.0960 | No |
| 50 | Rps6ka3 | 10939 | -0.026 | -0.1055 | No |
| 51 | Cdkn1a | 10951 | -0.026 | -0.1047 | No |
| 52 | Pfn1 | 10968 | -0.027 | -0.1041 | No |
| 53 | Map2k6 | 11247 | -0.037 | -0.1164 | No |
| 54 | Sla | 11317 | -0.040 | -0.1179 | No |
| 55 | Tnfrsf1a | 11353 | -0.041 | -0.1175 | No |
| 56 | Pdk1 | 11600 | -0.050 | -0.1276 | No |
| 57 | Mapkap1 | 11682 | -0.053 | -0.1290 | No |
| 58 | Camk4 | 11694 | -0.053 | -0.1267 | No |
| 59 | Akt1s1 | 11973 | -0.064 | -0.1376 | No |
| 60 | Ube2d3 | 12019 | -0.066 | -0.1364 | No |
| 61 | Nfkbib | 12151 | -0.072 | -0.1394 | No |
| 62 | Arpc3 | 12394 | -0.079 | -0.1476 | No |
| 63 | Cab39 | 13018 | -0.105 | -0.1742 | No |
| 64 | Pten | 13212 | -0.111 | -0.1782 | No |
| 65 | Them4 | 13271 | -0.114 | -0.1752 | No |
| 66 | Mapk10 | 13335 | -0.117 | -0.1723 | No |
| 67 | Rac1 | 13352 | -0.117 | -0.1669 | No |
| 68 | Grb2 | 13566 | -0.127 | -0.1712 | No |
| 69 | Cab39l | 13840 | -0.139 | -0.1779 | No |
| 70 | Cdk1 | 14093 | -0.150 | -0.1830 | Yes |
| 71 | Cltc | 14118 | -0.151 | -0.1762 | Yes |
| 72 | Arf1 | 14126 | -0.152 | -0.1686 | Yes |
| 73 | Il2rg | 14152 | -0.153 | -0.1618 | Yes |
| 74 | Dapp1 | 14291 | -0.160 | -0.1604 | Yes |
| 75 | Nck1 | 14510 | -0.170 | -0.1627 | Yes |
| 76 | Map3k7 | 14617 | -0.175 | -0.1589 | Yes |
| 77 | Atf1 | 14623 | -0.175 | -0.1499 | Yes |
| 78 | Dusp3 | 14659 | -0.177 | -0.1424 | Yes |
| 79 | Ecsit | 14886 | -0.189 | -0.1440 | Yes |
| 80 | Rps6ka1 | 15056 | -0.199 | -0.1422 | Yes |
| 81 | Stat2 | 15178 | -0.205 | -0.1376 | Yes |
| 82 | Irak4 | 15524 | -0.223 | -0.1436 | Yes |
| 83 | Actr3 | 15654 | -0.230 | -0.1381 | Yes |
| 84 | Akt1 | 15850 | -0.242 | -0.1354 | Yes |
| 85 | Ppp1ca | 16029 | -0.252 | -0.1312 | Yes |
| 86 | Traf2 | 16247 | -0.267 | -0.1283 | Yes |
| 87 | Myd88 | 16752 | -0.296 | -0.1386 | Yes |
| 88 | Calr | 17067 | -0.312 | -0.1383 | Yes |
| 89 | Cdkn1b | 17224 | -0.324 | -0.1292 | Yes |
| 90 | Mknk1 | 17392 | -0.333 | -0.1202 | Yes |
| 91 | Pik3r3 | 18455 | -0.455 | -0.1508 | Yes |
| 92 | Csnk2b | 18473 | -0.459 | -0.1274 | Yes |
| 93 | Rit1 | 18502 | -0.463 | -0.1044 | Yes |
| 94 | Gna14 | 18824 | -0.515 | -0.0937 | Yes |
| 95 | Cfl1 | 18879 | -0.524 | -0.0688 | Yes |
| 96 | Hras | 19031 | -0.565 | -0.0467 | Yes |
| 97 | Ddit3 | 19218 | -0.646 | -0.0222 | Yes |
| 98 | Slc2a1 | 19320 | -0.705 | 0.0099 | Yes |