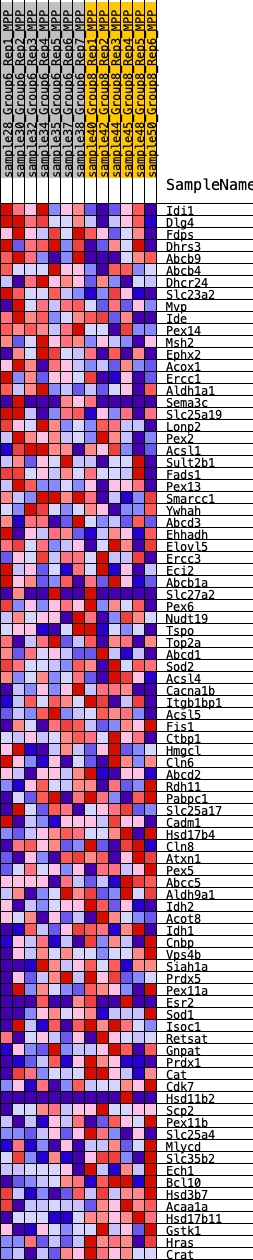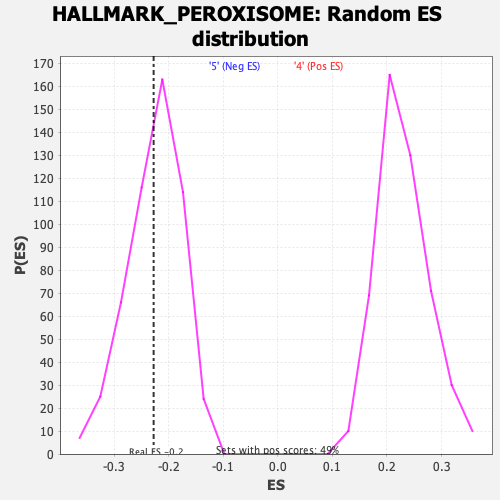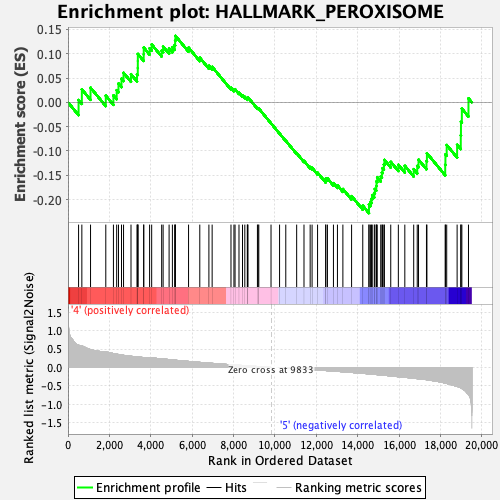
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group6_versus_Group8.MPP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | MPP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.22796234 |
| Normalized Enrichment Score (NES) | -1.0092816 |
| Nominal p-value | 0.44271845 |
| FDR q-value | 0.7942521 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Idi1 | 511 | 0.602 | 0.0048 | No |
| 2 | Dlg4 | 669 | 0.581 | 0.0268 | No |
| 3 | Fdps | 1089 | 0.483 | 0.0301 | No |
| 4 | Dhrs3 | 1826 | 0.416 | 0.0138 | No |
| 5 | Abcb9 | 2195 | 0.378 | 0.0143 | No |
| 6 | Abcb4 | 2352 | 0.361 | 0.0250 | No |
| 7 | Dhcr24 | 2439 | 0.352 | 0.0388 | No |
| 8 | Slc23a2 | 2585 | 0.338 | 0.0488 | No |
| 9 | Mvp | 2684 | 0.329 | 0.0608 | No |
| 10 | Ide | 3044 | 0.302 | 0.0579 | No |
| 11 | Pex14 | 3334 | 0.286 | 0.0578 | No |
| 12 | Msh2 | 3372 | 0.282 | 0.0705 | No |
| 13 | Ephx2 | 3374 | 0.282 | 0.0851 | No |
| 14 | Acox1 | 3375 | 0.282 | 0.0997 | No |
| 15 | Ercc1 | 3657 | 0.267 | 0.0990 | No |
| 16 | Aldh1a1 | 3660 | 0.267 | 0.1127 | No |
| 17 | Sema3c | 3943 | 0.253 | 0.1112 | No |
| 18 | Slc25a19 | 4043 | 0.252 | 0.1191 | No |
| 19 | Lonp2 | 4526 | 0.235 | 0.1065 | No |
| 20 | Pex2 | 4596 | 0.231 | 0.1149 | No |
| 21 | Acsl1 | 4885 | 0.214 | 0.1111 | No |
| 22 | Sult2b1 | 5044 | 0.204 | 0.1136 | No |
| 23 | Fads1 | 5156 | 0.198 | 0.1181 | No |
| 24 | Pex13 | 5178 | 0.197 | 0.1272 | No |
| 25 | Smarcc1 | 5190 | 0.197 | 0.1368 | No |
| 26 | Ywhah | 5825 | 0.164 | 0.1127 | No |
| 27 | Abcd3 | 6368 | 0.138 | 0.0919 | No |
| 28 | Ehhadh | 6808 | 0.118 | 0.0754 | No |
| 29 | Elovl5 | 6965 | 0.111 | 0.0731 | No |
| 30 | Ercc3 | 7875 | 0.073 | 0.0301 | No |
| 31 | Eci2 | 8018 | 0.068 | 0.0263 | No |
| 32 | Abcb1a | 8072 | 0.066 | 0.0270 | No |
| 33 | Slc27a2 | 8262 | 0.059 | 0.0203 | No |
| 34 | Pex6 | 8432 | 0.053 | 0.0144 | No |
| 35 | Nudt19 | 8543 | 0.049 | 0.0112 | No |
| 36 | Tspo | 8664 | 0.044 | 0.0073 | No |
| 37 | Top2a | 8677 | 0.044 | 0.0090 | No |
| 38 | Abcd1 | 8695 | 0.043 | 0.0103 | No |
| 39 | Sod2 | 9153 | 0.026 | -0.0119 | No |
| 40 | Acsl4 | 9184 | 0.024 | -0.0122 | No |
| 41 | Cacna1b | 9219 | 0.023 | -0.0128 | No |
| 42 | Itgb1bp1 | 9810 | 0.001 | -0.0431 | No |
| 43 | Acsl5 | 10223 | -0.002 | -0.0642 | No |
| 44 | Fis1 | 10523 | -0.011 | -0.0791 | No |
| 45 | Ctbp1 | 11049 | -0.030 | -0.1046 | No |
| 46 | Hmgcl | 11403 | -0.043 | -0.1205 | No |
| 47 | Cln6 | 11701 | -0.054 | -0.1330 | No |
| 48 | Abcd2 | 11794 | -0.057 | -0.1348 | No |
| 49 | Rdh11 | 12059 | -0.068 | -0.1448 | No |
| 50 | Pabpc1 | 12452 | -0.081 | -0.1608 | No |
| 51 | Slc25a17 | 12456 | -0.082 | -0.1567 | No |
| 52 | Cadm1 | 12536 | -0.084 | -0.1564 | No |
| 53 | Hsd17b4 | 12826 | -0.097 | -0.1663 | No |
| 54 | Cln8 | 13019 | -0.105 | -0.1707 | No |
| 55 | Atxn1 | 13282 | -0.114 | -0.1783 | No |
| 56 | Pex5 | 13703 | -0.134 | -0.1930 | No |
| 57 | Abcc5 | 14243 | -0.157 | -0.2126 | No |
| 58 | Aldh9a1 | 14542 | -0.171 | -0.2191 | Yes |
| 59 | Idh2 | 14543 | -0.172 | -0.2102 | Yes |
| 60 | Acot8 | 14612 | -0.174 | -0.2047 | Yes |
| 61 | Idh1 | 14667 | -0.177 | -0.1983 | Yes |
| 62 | Cnbp | 14700 | -0.178 | -0.1908 | Yes |
| 63 | Vps4b | 14800 | -0.184 | -0.1864 | Yes |
| 64 | Siah1a | 14827 | -0.185 | -0.1781 | Yes |
| 65 | Prdx5 | 14898 | -0.190 | -0.1719 | Yes |
| 66 | Pex11a | 14907 | -0.190 | -0.1625 | Yes |
| 67 | Esr2 | 14942 | -0.193 | -0.1543 | Yes |
| 68 | Sod1 | 15111 | -0.202 | -0.1525 | Yes |
| 69 | Isoc1 | 15168 | -0.205 | -0.1448 | Yes |
| 70 | Retsat | 15197 | -0.206 | -0.1356 | Yes |
| 71 | Gnpat | 15263 | -0.209 | -0.1281 | Yes |
| 72 | Prdx1 | 15288 | -0.211 | -0.1184 | Yes |
| 73 | Cat | 15595 | -0.227 | -0.1224 | Yes |
| 74 | Cdk7 | 15962 | -0.249 | -0.1284 | Yes |
| 75 | Hsd11b2 | 16275 | -0.269 | -0.1306 | Yes |
| 76 | Scp2 | 16704 | -0.293 | -0.1375 | Yes |
| 77 | Pex11b | 16875 | -0.302 | -0.1306 | Yes |
| 78 | Slc25a4 | 16937 | -0.307 | -0.1179 | Yes |
| 79 | Mlycd | 17321 | -0.329 | -0.1207 | Yes |
| 80 | Slc35b2 | 17343 | -0.330 | -0.1047 | Yes |
| 81 | Ech1 | 18227 | -0.421 | -0.1283 | Yes |
| 82 | Bcl10 | 18237 | -0.423 | -0.1069 | Yes |
| 83 | Hsd3b7 | 18299 | -0.431 | -0.0878 | Yes |
| 84 | Acaa1a | 18802 | -0.512 | -0.0871 | Yes |
| 85 | Hsd17b11 | 18979 | -0.547 | -0.0679 | Yes |
| 86 | Gstk1 | 18988 | -0.549 | -0.0399 | Yes |
| 87 | Hras | 19031 | -0.565 | -0.0129 | Yes |
| 88 | Crat | 19351 | -0.727 | 0.0083 | Yes |