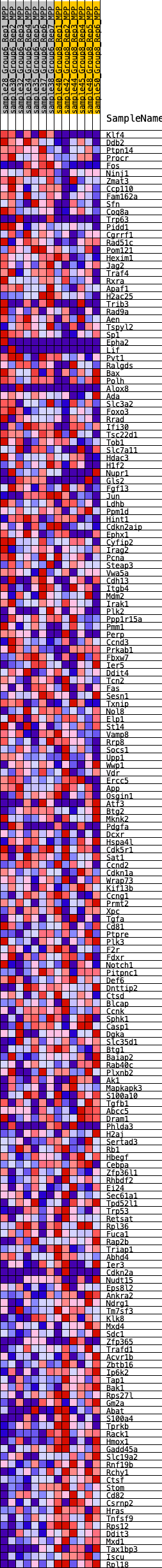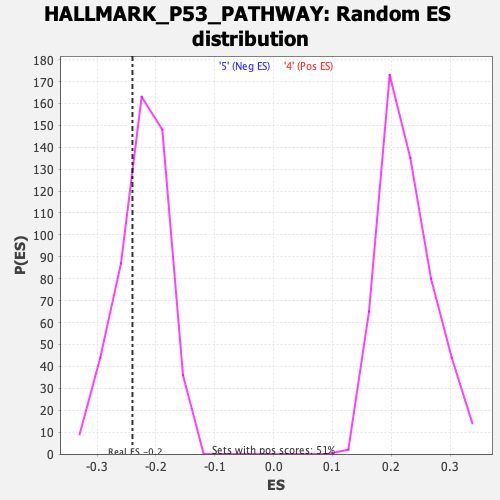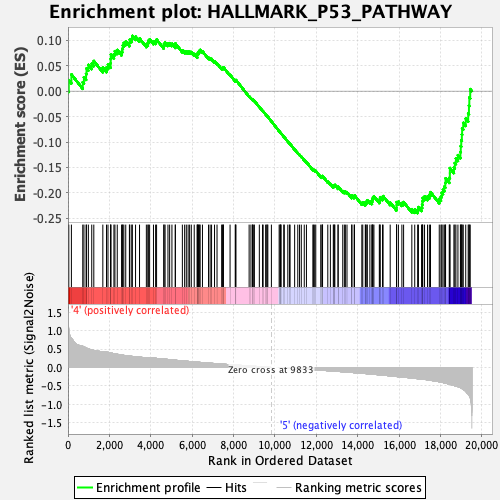
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group6_versus_Group8.MPP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | MPP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_P53_PATHWAY |
| Enrichment Score (ES) | -0.2397729 |
| Normalized Enrichment Score (NES) | -1.0771941 |
| Nominal p-value | 0.29568788 |
| FDR q-value | 0.630893 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Klf4 | 38 | 1.039 | 0.0216 | No |
| 2 | Ddb2 | 164 | 0.784 | 0.0329 | No |
| 3 | Ptpn14 | 713 | 0.575 | 0.0175 | No |
| 4 | Procr | 770 | 0.560 | 0.0273 | No |
| 5 | Fos | 874 | 0.530 | 0.0340 | No |
| 6 | Ninj1 | 896 | 0.524 | 0.0448 | No |
| 7 | Zmat3 | 984 | 0.502 | 0.0516 | No |
| 8 | Ccp110 | 1147 | 0.474 | 0.0540 | No |
| 9 | Fam162a | 1247 | 0.461 | 0.0593 | No |
| 10 | Sfn | 1683 | 0.423 | 0.0464 | No |
| 11 | Coq8a | 1860 | 0.413 | 0.0467 | No |
| 12 | Trp63 | 1932 | 0.409 | 0.0522 | No |
| 13 | Pidd1 | 2057 | 0.393 | 0.0547 | No |
| 14 | Cgrrf1 | 2063 | 0.392 | 0.0634 | No |
| 15 | Rad51c | 2073 | 0.391 | 0.0717 | No |
| 16 | Pom121 | 2219 | 0.374 | 0.0727 | No |
| 17 | Hexim1 | 2267 | 0.369 | 0.0787 | No |
| 18 | Jag2 | 2380 | 0.358 | 0.0810 | No |
| 19 | Traf4 | 2589 | 0.337 | 0.0778 | No |
| 20 | Rxra | 2626 | 0.334 | 0.0836 | No |
| 21 | Apaf1 | 2649 | 0.332 | 0.0899 | No |
| 22 | H2ac25 | 2698 | 0.328 | 0.0949 | No |
| 23 | Trib3 | 2790 | 0.320 | 0.0974 | No |
| 24 | Rad9a | 2967 | 0.307 | 0.0953 | No |
| 25 | Aen | 2987 | 0.306 | 0.1012 | No |
| 26 | Tspyl2 | 3082 | 0.301 | 0.1032 | No |
| 27 | Sp1 | 3108 | 0.299 | 0.1086 | No |
| 28 | Epha2 | 3264 | 0.289 | 0.1072 | No |
| 29 | Lif | 3456 | 0.277 | 0.1036 | No |
| 30 | Pvt1 | 3786 | 0.263 | 0.0925 | No |
| 31 | Ralgds | 3859 | 0.258 | 0.0946 | No |
| 32 | Bax | 3881 | 0.256 | 0.0993 | No |
| 33 | Polh | 3944 | 0.253 | 0.1018 | No |
| 34 | Alox8 | 4130 | 0.251 | 0.0980 | No |
| 35 | Ada | 4234 | 0.248 | 0.0982 | No |
| 36 | Slc3a2 | 4280 | 0.245 | 0.1015 | No |
| 37 | Foxo3 | 4625 | 0.229 | 0.0889 | No |
| 38 | Rrad | 4643 | 0.228 | 0.0931 | No |
| 39 | Ifi30 | 4691 | 0.225 | 0.0958 | No |
| 40 | Tsc22d1 | 4830 | 0.217 | 0.0936 | No |
| 41 | Tob1 | 4911 | 0.212 | 0.0942 | No |
| 42 | Slc7a11 | 5022 | 0.206 | 0.0932 | No |
| 43 | Hdac3 | 5185 | 0.197 | 0.0893 | No |
| 44 | H1f2 | 5186 | 0.197 | 0.0937 | No |
| 45 | Nupr1 | 5526 | 0.178 | 0.0803 | No |
| 46 | Gls2 | 5636 | 0.173 | 0.0785 | No |
| 47 | Fgf13 | 5716 | 0.170 | 0.0783 | No |
| 48 | Jun | 5804 | 0.164 | 0.0775 | No |
| 49 | Ldhb | 5877 | 0.161 | 0.0774 | No |
| 50 | Ppm1d | 5958 | 0.158 | 0.0769 | No |
| 51 | Hint1 | 6099 | 0.151 | 0.0731 | No |
| 52 | Cdkn2aip | 6245 | 0.144 | 0.0688 | No |
| 53 | Ephx1 | 6246 | 0.144 | 0.0721 | No |
| 54 | Cyfip2 | 6263 | 0.143 | 0.0745 | No |
| 55 | Irag2 | 6304 | 0.141 | 0.0756 | No |
| 56 | Pcna | 6321 | 0.140 | 0.0779 | No |
| 57 | Steap3 | 6362 | 0.138 | 0.0790 | No |
| 58 | Vwa5a | 6382 | 0.137 | 0.0811 | No |
| 59 | Cdh13 | 6497 | 0.132 | 0.0782 | No |
| 60 | Itgb4 | 6791 | 0.119 | 0.0657 | No |
| 61 | Mdm2 | 6867 | 0.115 | 0.0644 | No |
| 62 | Irak1 | 6937 | 0.112 | 0.0634 | No |
| 63 | Plk2 | 7080 | 0.106 | 0.0585 | No |
| 64 | Ppp1r15a | 7201 | 0.101 | 0.0546 | No |
| 65 | Pmm1 | 7425 | 0.091 | 0.0451 | No |
| 66 | Perp | 7462 | 0.090 | 0.0452 | No |
| 67 | Ccnd3 | 7503 | 0.088 | 0.0452 | No |
| 68 | Prkab1 | 7507 | 0.088 | 0.0470 | No |
| 69 | Fbxw7 | 7831 | 0.076 | 0.0320 | No |
| 70 | Ier5 | 8084 | 0.065 | 0.0204 | No |
| 71 | Ddit4 | 8089 | 0.065 | 0.0217 | No |
| 72 | Tcn2 | 8116 | 0.064 | 0.0218 | No |
| 73 | Fas | 8752 | 0.041 | -0.0101 | No |
| 74 | Sesn1 | 8840 | 0.037 | -0.0138 | No |
| 75 | Txnip | 8915 | 0.035 | -0.0168 | No |
| 76 | Nol8 | 8917 | 0.035 | -0.0161 | No |
| 77 | Elp1 | 8973 | 0.032 | -0.0182 | No |
| 78 | St14 | 9005 | 0.031 | -0.0191 | No |
| 79 | Vamp8 | 9245 | 0.022 | -0.0310 | No |
| 80 | Rrp8 | 9399 | 0.016 | -0.0385 | No |
| 81 | Socs1 | 9428 | 0.015 | -0.0396 | No |
| 82 | Upp1 | 9550 | 0.010 | -0.0457 | No |
| 83 | Wwp1 | 9609 | 0.008 | -0.0485 | No |
| 84 | Vdr | 9617 | 0.008 | -0.0487 | No |
| 85 | Ercc5 | 9656 | 0.006 | -0.0505 | No |
| 86 | App | 9828 | 0.000 | -0.0593 | No |
| 87 | Osgin1 | 10203 | -0.001 | -0.0787 | No |
| 88 | Atf3 | 10243 | -0.003 | -0.0806 | No |
| 89 | Btg2 | 10271 | -0.003 | -0.0819 | No |
| 90 | Mknk2 | 10276 | -0.004 | -0.0821 | No |
| 91 | Pdgfa | 10294 | -0.005 | -0.0829 | No |
| 92 | Dcxr | 10426 | -0.008 | -0.0894 | No |
| 93 | Hspa4l | 10450 | -0.009 | -0.0904 | No |
| 94 | Cdk5r1 | 10621 | -0.014 | -0.0989 | No |
| 95 | Sat1 | 10700 | -0.016 | -0.1026 | No |
| 96 | Ccnd2 | 10731 | -0.017 | -0.1038 | No |
| 97 | Cdkn1a | 10951 | -0.026 | -0.1145 | No |
| 98 | Wrap73 | 11103 | -0.032 | -0.1216 | No |
| 99 | Kif13b | 11188 | -0.035 | -0.1251 | No |
| 100 | Ccng1 | 11273 | -0.038 | -0.1286 | No |
| 101 | Prmt2 | 11416 | -0.044 | -0.1350 | No |
| 102 | Xpc | 11521 | -0.047 | -0.1393 | No |
| 103 | Tgfa | 11832 | -0.058 | -0.1540 | No |
| 104 | Cd81 | 11867 | -0.060 | -0.1544 | No |
| 105 | Ptpre | 11919 | -0.062 | -0.1556 | No |
| 106 | Plk3 | 11972 | -0.064 | -0.1569 | No |
| 107 | F2r | 12212 | -0.075 | -0.1675 | No |
| 108 | Fdxr | 12267 | -0.076 | -0.1686 | No |
| 109 | Notch1 | 12296 | -0.077 | -0.1683 | No |
| 110 | Pitpnc1 | 12300 | -0.077 | -0.1667 | No |
| 111 | Def6 | 12553 | -0.086 | -0.1778 | No |
| 112 | Dnttip2 | 12678 | -0.091 | -0.1822 | No |
| 113 | Ctsd | 12819 | -0.097 | -0.1872 | No |
| 114 | Blcap | 12849 | -0.098 | -0.1865 | No |
| 115 | Ccnk | 12867 | -0.099 | -0.1851 | No |
| 116 | Sphk1 | 12894 | -0.100 | -0.1842 | No |
| 117 | Casp1 | 13050 | -0.106 | -0.1898 | No |
| 118 | Dgka | 13051 | -0.106 | -0.1874 | No |
| 119 | Slc35d1 | 13276 | -0.114 | -0.1964 | No |
| 120 | Btg1 | 13354 | -0.117 | -0.1978 | No |
| 121 | Baiap2 | 13402 | -0.120 | -0.1975 | No |
| 122 | Rab40c | 13481 | -0.123 | -0.1987 | No |
| 123 | Plxnb2 | 13707 | -0.134 | -0.2074 | No |
| 124 | Ak1 | 13720 | -0.134 | -0.2049 | No |
| 125 | Mapkapk3 | 13827 | -0.138 | -0.2073 | No |
| 126 | S100a10 | 13844 | -0.139 | -0.2050 | No |
| 127 | Tgfb1 | 14198 | -0.155 | -0.2197 | No |
| 128 | Abcc5 | 14243 | -0.157 | -0.2184 | No |
| 129 | Dram1 | 14365 | -0.163 | -0.2210 | No |
| 130 | Phlda3 | 14378 | -0.164 | -0.2179 | No |
| 131 | H2aj | 14438 | -0.167 | -0.2172 | No |
| 132 | Sertad3 | 14460 | -0.168 | -0.2145 | No |
| 133 | Rb1 | 14587 | -0.174 | -0.2170 | No |
| 134 | Hbegf | 14685 | -0.178 | -0.2180 | No |
| 135 | Cebpa | 14695 | -0.178 | -0.2145 | No |
| 136 | Zfp36l1 | 14715 | -0.179 | -0.2114 | No |
| 137 | Rhbdf2 | 14738 | -0.180 | -0.2085 | No |
| 138 | Ei24 | 14782 | -0.182 | -0.2066 | No |
| 139 | Sec61a1 | 15043 | -0.198 | -0.2155 | No |
| 140 | Tpd52l1 | 15068 | -0.200 | -0.2122 | No |
| 141 | Trp53 | 15084 | -0.201 | -0.2085 | No |
| 142 | Retsat | 15197 | -0.206 | -0.2096 | No |
| 143 | Rpl36 | 15225 | -0.208 | -0.2063 | No |
| 144 | Fuca1 | 15568 | -0.225 | -0.2189 | No |
| 145 | Rap2b | 15873 | -0.243 | -0.2291 | No |
| 146 | Triap1 | 15876 | -0.244 | -0.2237 | No |
| 147 | Abhd4 | 15877 | -0.244 | -0.2181 | No |
| 148 | Ier3 | 15965 | -0.249 | -0.2170 | No |
| 149 | Cdkn2a | 16133 | -0.260 | -0.2198 | No |
| 150 | Nudt15 | 16216 | -0.265 | -0.2180 | No |
| 151 | Eps8l2 | 16614 | -0.287 | -0.2320 | No |
| 152 | Ankra2 | 16754 | -0.296 | -0.2325 | No |
| 153 | Ndrg1 | 16895 | -0.304 | -0.2329 | Yes |
| 154 | Tm7sf3 | 16933 | -0.306 | -0.2279 | Yes |
| 155 | Klk8 | 17085 | -0.314 | -0.2286 | Yes |
| 156 | Mxd4 | 17112 | -0.316 | -0.2228 | Yes |
| 157 | Sdc1 | 17129 | -0.317 | -0.2164 | Yes |
| 158 | Zfp365 | 17142 | -0.319 | -0.2098 | Yes |
| 159 | Trafd1 | 17228 | -0.325 | -0.2069 | Yes |
| 160 | Acvr1b | 17373 | -0.332 | -0.2068 | Yes |
| 161 | Zbtb16 | 17461 | -0.340 | -0.2036 | Yes |
| 162 | Ip6k2 | 17516 | -0.346 | -0.1985 | Yes |
| 163 | Tap1 | 17939 | -0.385 | -0.2116 | Yes |
| 164 | Bak1 | 18021 | -0.395 | -0.2069 | Yes |
| 165 | Rps27l | 18060 | -0.400 | -0.1998 | Yes |
| 166 | Gm2a | 18119 | -0.407 | -0.1936 | Yes |
| 167 | Abat | 18184 | -0.416 | -0.1875 | Yes |
| 168 | S100a4 | 18232 | -0.422 | -0.1803 | Yes |
| 169 | Tprkb | 18244 | -0.423 | -0.1713 | Yes |
| 170 | Rack1 | 18422 | -0.450 | -0.1702 | Yes |
| 171 | Hmox1 | 18451 | -0.455 | -0.1614 | Yes |
| 172 | Gadd45a | 18452 | -0.455 | -0.1511 | Yes |
| 173 | Slc19a2 | 18647 | -0.484 | -0.1501 | Yes |
| 174 | Rnf19b | 18691 | -0.492 | -0.1412 | Yes |
| 175 | Rchy1 | 18743 | -0.503 | -0.1325 | Yes |
| 176 | Ctsf | 18841 | -0.518 | -0.1257 | Yes |
| 177 | Stom | 18961 | -0.544 | -0.1196 | Yes |
| 178 | Cd82 | 18978 | -0.547 | -0.1080 | Yes |
| 179 | Csrnp2 | 19002 | -0.556 | -0.0966 | Yes |
| 180 | Hras | 19031 | -0.565 | -0.0853 | Yes |
| 181 | Tnfsf9 | 19044 | -0.569 | -0.0730 | Yes |
| 182 | Rps12 | 19098 | -0.592 | -0.0623 | Yes |
| 183 | Ddit3 | 19218 | -0.646 | -0.0538 | Yes |
| 184 | Mxd1 | 19337 | -0.715 | -0.0437 | Yes |
| 185 | Tax1bp3 | 19374 | -0.745 | -0.0287 | Yes |
| 186 | Iscu | 19392 | -0.764 | -0.0123 | Yes |
| 187 | Rpl18 | 19435 | -0.815 | 0.0040 | Yes |