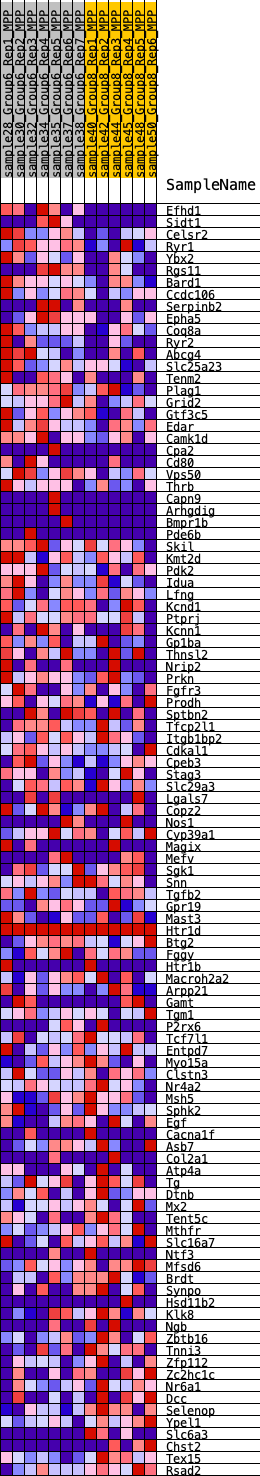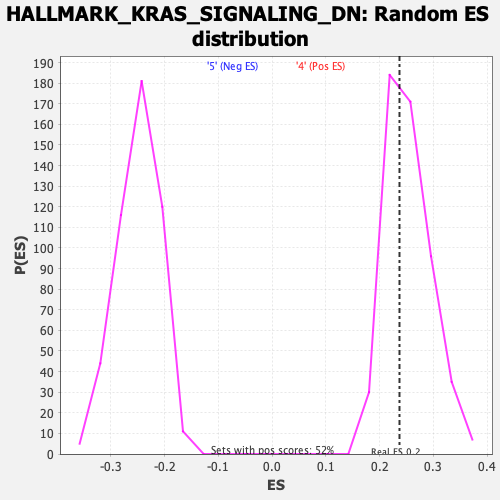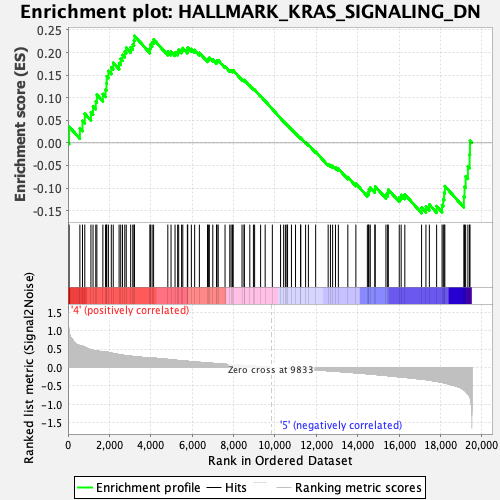
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group6_versus_Group8.MPP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | MPP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.23686422 |
| Normalized Enrichment Score (NES) | 0.9388334 |
| Nominal p-value | 0.6003824 |
| FDR q-value | 0.931191 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Efhd1 | 53 | 0.988 | 0.0360 | Yes |
| 2 | Sidt1 | 574 | 0.594 | 0.0325 | Yes |
| 3 | Celsr2 | 700 | 0.578 | 0.0487 | Yes |
| 4 | Ryr1 | 811 | 0.547 | 0.0645 | Yes |
| 5 | Ybx2 | 1108 | 0.480 | 0.0681 | Yes |
| 6 | Rgs11 | 1210 | 0.465 | 0.0811 | Yes |
| 7 | Bard1 | 1342 | 0.448 | 0.0919 | Yes |
| 8 | Ccdc106 | 1395 | 0.441 | 0.1066 | Yes |
| 9 | Serpinb2 | 1685 | 0.423 | 0.1083 | Yes |
| 10 | Epha5 | 1814 | 0.418 | 0.1180 | Yes |
| 11 | Coq8a | 1860 | 0.413 | 0.1319 | Yes |
| 12 | Ryr2 | 1872 | 0.411 | 0.1475 | Yes |
| 13 | Abcg4 | 1952 | 0.409 | 0.1594 | Yes |
| 14 | Slc25a23 | 2092 | 0.389 | 0.1675 | Yes |
| 15 | Tenm2 | 2186 | 0.379 | 0.1776 | Yes |
| 16 | Plag1 | 2469 | 0.349 | 0.1767 | Yes |
| 17 | Grid2 | 2548 | 0.342 | 0.1861 | Yes |
| 18 | Gtf3c5 | 2634 | 0.334 | 0.1948 | Yes |
| 19 | Edar | 2744 | 0.323 | 0.2019 | Yes |
| 20 | Camk1d | 2814 | 0.319 | 0.2108 | Yes |
| 21 | Cpa2 | 3028 | 0.303 | 0.2117 | Yes |
| 22 | Cd80 | 3131 | 0.297 | 0.2181 | Yes |
| 23 | Vps50 | 3187 | 0.293 | 0.2268 | Yes |
| 24 | Thrb | 3214 | 0.292 | 0.2369 | Yes |
| 25 | Capn9 | 3959 | 0.252 | 0.2084 | No |
| 26 | Arhgdig | 3978 | 0.252 | 0.2174 | No |
| 27 | Bmpr1b | 4064 | 0.251 | 0.2229 | No |
| 28 | Pde6b | 4136 | 0.251 | 0.2290 | No |
| 29 | Skil | 4823 | 0.217 | 0.2022 | No |
| 30 | Kmt2d | 4979 | 0.208 | 0.2024 | No |
| 31 | Pdk2 | 5172 | 0.198 | 0.2002 | No |
| 32 | Idua | 5298 | 0.189 | 0.2012 | No |
| 33 | Lfng | 5348 | 0.187 | 0.2060 | No |
| 34 | Kcnd1 | 5488 | 0.180 | 0.2059 | No |
| 35 | Ptprj | 5547 | 0.177 | 0.2098 | No |
| 36 | Kcnn1 | 5767 | 0.167 | 0.2051 | No |
| 37 | Gp1ba | 5785 | 0.166 | 0.2107 | No |
| 38 | Thnsl2 | 5961 | 0.158 | 0.2079 | No |
| 39 | Nrip2 | 6120 | 0.150 | 0.2056 | No |
| 40 | Prkn | 6348 | 0.139 | 0.1994 | No |
| 41 | Fgfr3 | 6741 | 0.120 | 0.1839 | No |
| 42 | Prodh | 6784 | 0.119 | 0.1864 | No |
| 43 | Sptbn2 | 6829 | 0.117 | 0.1887 | No |
| 44 | Tfcp2l1 | 6996 | 0.110 | 0.1845 | No |
| 45 | Itgb1bp2 | 7175 | 0.102 | 0.1793 | No |
| 46 | Cdkal1 | 7200 | 0.101 | 0.1820 | No |
| 47 | Cpeb3 | 7258 | 0.098 | 0.1829 | No |
| 48 | Stag3 | 7588 | 0.085 | 0.1693 | No |
| 49 | Slc29a3 | 7818 | 0.076 | 0.1605 | No |
| 50 | Lgals7 | 7882 | 0.073 | 0.1601 | No |
| 51 | Copz2 | 7951 | 0.071 | 0.1594 | No |
| 52 | Nos1 | 7982 | 0.070 | 0.1606 | No |
| 53 | Cyp39a1 | 8409 | 0.054 | 0.1407 | No |
| 54 | Magix | 8507 | 0.050 | 0.1377 | No |
| 55 | Mefv | 8519 | 0.050 | 0.1391 | No |
| 56 | Sgk1 | 8785 | 0.040 | 0.1270 | No |
| 57 | Snn | 8959 | 0.033 | 0.1193 | No |
| 58 | Tgfb2 | 9017 | 0.030 | 0.1176 | No |
| 59 | Gpr19 | 9307 | 0.020 | 0.1035 | No |
| 60 | Mast3 | 9527 | 0.011 | 0.0926 | No |
| 61 | Htr1d | 9874 | 0.000 | 0.0748 | No |
| 62 | Btg2 | 10271 | -0.003 | 0.0545 | No |
| 63 | Fggy | 10420 | -0.008 | 0.0472 | No |
| 64 | Htr1b | 10505 | -0.010 | 0.0433 | No |
| 65 | Macroh2a2 | 10578 | -0.012 | 0.0401 | No |
| 66 | Arpp21 | 10609 | -0.013 | 0.0390 | No |
| 67 | Gamt | 10797 | -0.020 | 0.0302 | No |
| 68 | Tgm1 | 10994 | -0.028 | 0.0211 | No |
| 69 | P2rx6 | 11238 | -0.037 | 0.0101 | No |
| 70 | Tcf7l1 | 11246 | -0.037 | 0.0112 | No |
| 71 | Entpd7 | 11480 | -0.045 | 0.0009 | No |
| 72 | Myo15a | 11616 | -0.050 | -0.0041 | No |
| 73 | Clstn3 | 11967 | -0.064 | -0.0196 | No |
| 74 | Nr4a2 | 12571 | -0.087 | -0.0473 | No |
| 75 | Msh5 | 12681 | -0.091 | -0.0493 | No |
| 76 | Sphk2 | 12787 | -0.096 | -0.0510 | No |
| 77 | Egf | 12925 | -0.102 | -0.0541 | No |
| 78 | Cacna1f | 13064 | -0.107 | -0.0570 | No |
| 79 | Asb7 | 13521 | -0.125 | -0.0756 | No |
| 80 | Col2a1 | 13910 | -0.142 | -0.0900 | No |
| 81 | Atp4a | 14465 | -0.168 | -0.1120 | No |
| 82 | Tg | 14524 | -0.171 | -0.1083 | No |
| 83 | Dtnb | 14541 | -0.171 | -0.1024 | No |
| 84 | Mx2 | 14606 | -0.174 | -0.0989 | No |
| 85 | Tent5c | 14826 | -0.185 | -0.1029 | No |
| 86 | Mthfr | 14843 | -0.186 | -0.0964 | No |
| 87 | Slc16a7 | 15363 | -0.215 | -0.1147 | No |
| 88 | Ntf3 | 15446 | -0.219 | -0.1104 | No |
| 89 | Mfsd6 | 15476 | -0.221 | -0.1032 | No |
| 90 | Brdt | 16006 | -0.251 | -0.1207 | No |
| 91 | Synpo | 16098 | -0.257 | -0.1153 | No |
| 92 | Hsd11b2 | 16275 | -0.269 | -0.1138 | No |
| 93 | Klk8 | 17085 | -0.314 | -0.1432 | No |
| 94 | Ngb | 17295 | -0.327 | -0.1412 | No |
| 95 | Zbtb16 | 17461 | -0.340 | -0.1363 | No |
| 96 | Tnni3 | 17812 | -0.371 | -0.1398 | No |
| 97 | Zfp112 | 18075 | -0.402 | -0.1376 | No |
| 98 | Zc2hc1c | 18132 | -0.409 | -0.1244 | No |
| 99 | Nr6a1 | 18175 | -0.415 | -0.1103 | No |
| 100 | Dcc | 18209 | -0.419 | -0.0956 | No |
| 101 | Selenop | 19127 | -0.601 | -0.1193 | No |
| 102 | Ypel1 | 19161 | -0.617 | -0.0968 | No |
| 103 | Slc6a3 | 19213 | -0.643 | -0.0742 | No |
| 104 | Chst2 | 19323 | -0.706 | -0.0521 | No |
| 105 | Tex15 | 19404 | -0.774 | -0.0259 | No |
| 106 | Rsad2 | 19423 | -0.800 | 0.0046 | No |