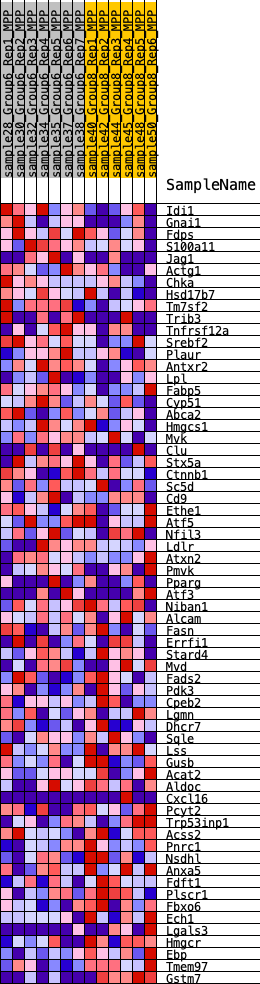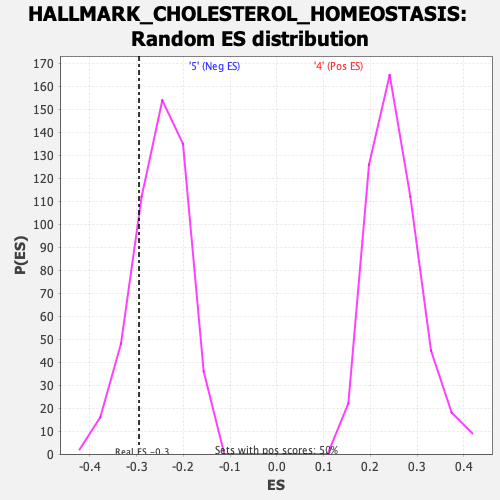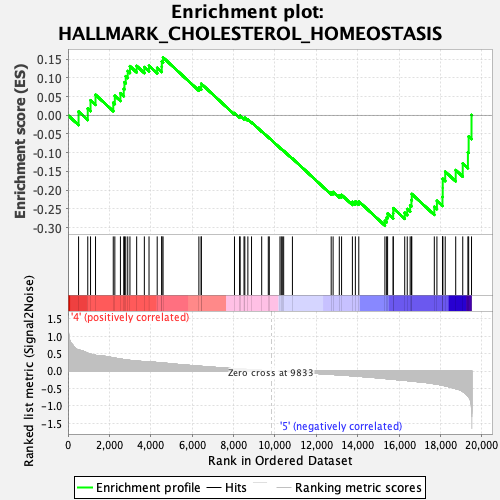
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group6_versus_Group8.MPP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | MPP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.29485497 |
| Normalized Enrichment Score (NES) | -1.1805449 |
| Nominal p-value | 0.19284295 |
| FDR q-value | 0.4401196 |
| FWER p-Value | 0.961 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Idi1 | 511 | 0.602 | 0.0099 | No |
| 2 | Gnai1 | 955 | 0.509 | 0.0178 | No |
| 3 | Fdps | 1089 | 0.483 | 0.0400 | No |
| 4 | S100a11 | 1330 | 0.450 | 0.0547 | No |
| 5 | Jag1 | 2187 | 0.379 | 0.0335 | No |
| 6 | Actg1 | 2263 | 0.370 | 0.0519 | No |
| 7 | Chka | 2533 | 0.344 | 0.0587 | No |
| 8 | Hsd17b7 | 2694 | 0.328 | 0.0703 | No |
| 9 | Tm7sf2 | 2728 | 0.325 | 0.0881 | No |
| 10 | Trib3 | 2790 | 0.320 | 0.1042 | No |
| 11 | Tnfrsf12a | 2887 | 0.313 | 0.1181 | No |
| 12 | Srebf2 | 2993 | 0.305 | 0.1311 | No |
| 13 | Plaur | 3318 | 0.288 | 0.1317 | No |
| 14 | Antxr2 | 3689 | 0.265 | 0.1286 | No |
| 15 | Lpl | 3915 | 0.254 | 0.1323 | No |
| 16 | Fabp5 | 4310 | 0.243 | 0.1267 | No |
| 17 | Cyp51 | 4530 | 0.235 | 0.1296 | No |
| 18 | Abca2 | 4534 | 0.235 | 0.1435 | No |
| 19 | Hmgcs1 | 4593 | 0.231 | 0.1545 | No |
| 20 | Mvk | 6324 | 0.140 | 0.0739 | No |
| 21 | Clu | 6437 | 0.135 | 0.0762 | No |
| 22 | Stx5a | 6438 | 0.135 | 0.0843 | No |
| 23 | Ctnnb1 | 8044 | 0.067 | 0.0058 | No |
| 24 | Sc5d | 8291 | 0.058 | -0.0033 | No |
| 25 | Cd9 | 8318 | 0.057 | -0.0013 | No |
| 26 | Ethe1 | 8505 | 0.050 | -0.0078 | No |
| 27 | Atf5 | 8544 | 0.049 | -0.0068 | No |
| 28 | Nfil3 | 8691 | 0.043 | -0.0117 | No |
| 29 | Ldlr | 8868 | 0.036 | -0.0186 | No |
| 30 | Atxn2 | 9359 | 0.018 | -0.0427 | No |
| 31 | Pmvk | 9678 | 0.005 | -0.0588 | No |
| 32 | Pparg | 9726 | 0.004 | -0.0610 | No |
| 33 | Atf3 | 10243 | -0.003 | -0.0873 | No |
| 34 | Niban1 | 10306 | -0.005 | -0.0902 | No |
| 35 | Alcam | 10355 | -0.007 | -0.0923 | No |
| 36 | Fasn | 10421 | -0.008 | -0.0951 | No |
| 37 | Errfi1 | 10844 | -0.022 | -0.1155 | No |
| 38 | Stard4 | 12717 | -0.092 | -0.2062 | No |
| 39 | Mvd | 12810 | -0.097 | -0.2051 | No |
| 40 | Fads2 | 13109 | -0.108 | -0.2140 | No |
| 41 | Pdk3 | 13219 | -0.112 | -0.2129 | No |
| 42 | Cpeb2 | 13742 | -0.135 | -0.2316 | No |
| 43 | Lgmn | 13885 | -0.141 | -0.2304 | No |
| 44 | Dhcr7 | 14051 | -0.147 | -0.2300 | No |
| 45 | Sqle | 15313 | -0.212 | -0.2821 | Yes |
| 46 | Lss | 15396 | -0.216 | -0.2733 | Yes |
| 47 | Gusb | 15443 | -0.219 | -0.2625 | Yes |
| 48 | Acat2 | 15711 | -0.234 | -0.2622 | Yes |
| 49 | Aldoc | 15716 | -0.234 | -0.2483 | Yes |
| 50 | Cxcl16 | 16270 | -0.269 | -0.2606 | Yes |
| 51 | Pcyt2 | 16392 | -0.271 | -0.2505 | Yes |
| 52 | Trp53inp1 | 16533 | -0.281 | -0.2408 | Yes |
| 53 | Acss2 | 16594 | -0.286 | -0.2267 | Yes |
| 54 | Pnrc1 | 16612 | -0.287 | -0.2104 | Yes |
| 55 | Nsdhl | 17700 | -0.361 | -0.2445 | Yes |
| 56 | Anxa5 | 17826 | -0.371 | -0.2286 | Yes |
| 57 | Fdft1 | 18095 | -0.404 | -0.2181 | Yes |
| 58 | Plscr1 | 18106 | -0.405 | -0.1943 | Yes |
| 59 | Fbxo6 | 18107 | -0.405 | -0.1699 | Yes |
| 60 | Ech1 | 18227 | -0.421 | -0.1506 | Yes |
| 61 | Lgals3 | 18734 | -0.501 | -0.1465 | Yes |
| 62 | Hmgcr | 19074 | -0.584 | -0.1288 | Yes |
| 63 | Ebp | 19324 | -0.708 | -0.0990 | Yes |
| 64 | Tmem97 | 19360 | -0.733 | -0.0567 | Yes |
| 65 | Gstm7 | 19498 | -1.072 | 0.0007 | Yes |