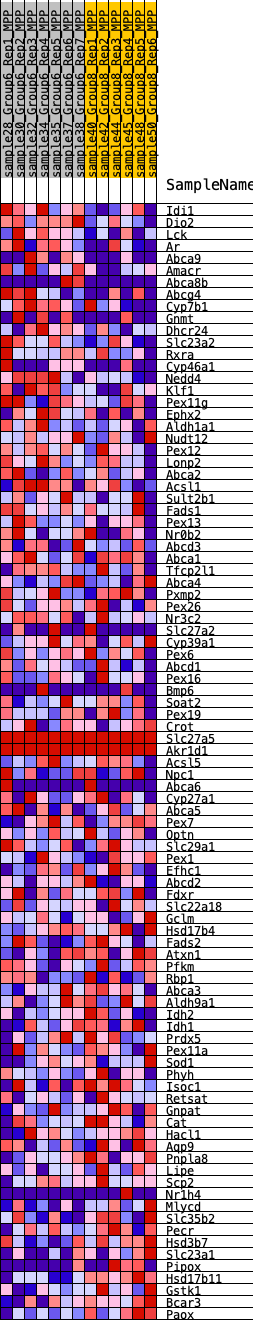
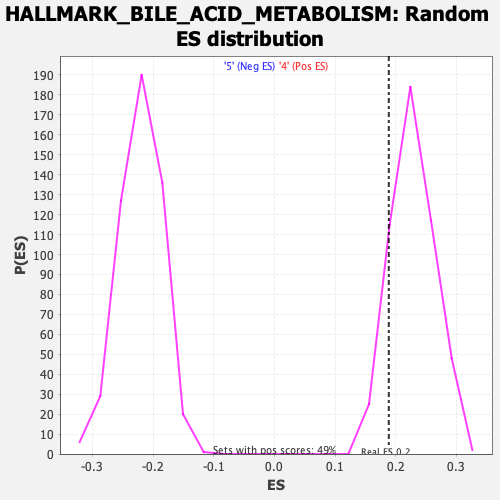
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group6_versus_Group8.MPP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | MPP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.18843101 |
| Normalized Enrichment Score (NES) | 0.8238028 |
| Nominal p-value | 0.8879837 |
| FDR q-value | 0.9378925 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Idi1 | 511 | 0.602 | 0.0031 | Yes |
| 2 | Dio2 | 742 | 0.566 | 0.0189 | Yes |
| 3 | Lck | 777 | 0.558 | 0.0444 | Yes |
| 4 | Ar | 1016 | 0.497 | 0.0564 | Yes |
| 5 | Abca9 | 1050 | 0.491 | 0.0787 | Yes |
| 6 | Amacr | 1072 | 0.486 | 0.1014 | Yes |
| 7 | Abca8b | 1909 | 0.409 | 0.0783 | Yes |
| 8 | Abcg4 | 1952 | 0.409 | 0.0961 | Yes |
| 9 | Cyp7b1 | 2221 | 0.374 | 0.1006 | Yes |
| 10 | Gnmt | 2417 | 0.354 | 0.1078 | Yes |
| 11 | Dhcr24 | 2439 | 0.352 | 0.1239 | Yes |
| 12 | Slc23a2 | 2585 | 0.338 | 0.1330 | Yes |
| 13 | Rxra | 2626 | 0.334 | 0.1472 | Yes |
| 14 | Cyp46a1 | 3018 | 0.303 | 0.1419 | Yes |
| 15 | Nedd4 | 3041 | 0.302 | 0.1555 | Yes |
| 16 | Klf1 | 3086 | 0.300 | 0.1679 | Yes |
| 17 | Pex11g | 3263 | 0.289 | 0.1730 | Yes |
| 18 | Ephx2 | 3374 | 0.282 | 0.1811 | Yes |
| 19 | Aldh1a1 | 3660 | 0.267 | 0.1795 | Yes |
| 20 | Nudt12 | 4244 | 0.248 | 0.1616 | Yes |
| 21 | Pex12 | 4305 | 0.244 | 0.1704 | Yes |
| 22 | Lonp2 | 4526 | 0.235 | 0.1705 | Yes |
| 23 | Abca2 | 4534 | 0.235 | 0.1816 | Yes |
| 24 | Acsl1 | 4885 | 0.214 | 0.1741 | Yes |
| 25 | Sult2b1 | 5044 | 0.204 | 0.1759 | Yes |
| 26 | Fads1 | 5156 | 0.198 | 0.1799 | Yes |
| 27 | Pex13 | 5178 | 0.197 | 0.1884 | Yes |
| 28 | Nr0b2 | 6299 | 0.141 | 0.1376 | No |
| 29 | Abcd3 | 6368 | 0.138 | 0.1409 | No |
| 30 | Abca1 | 6565 | 0.129 | 0.1371 | No |
| 31 | Tfcp2l1 | 6996 | 0.110 | 0.1203 | No |
| 32 | Abca4 | 7415 | 0.091 | 0.1032 | No |
| 33 | Pxmp2 | 8142 | 0.063 | 0.0689 | No |
| 34 | Pex26 | 8188 | 0.061 | 0.0696 | No |
| 35 | Nr3c2 | 8202 | 0.061 | 0.0719 | No |
| 36 | Slc27a2 | 8262 | 0.059 | 0.0717 | No |
| 37 | Cyp39a1 | 8409 | 0.054 | 0.0668 | No |
| 38 | Pex6 | 8432 | 0.053 | 0.0683 | No |
| 39 | Abcd1 | 8695 | 0.043 | 0.0569 | No |
| 40 | Pex16 | 8936 | 0.034 | 0.0462 | No |
| 41 | Bmp6 | 9287 | 0.020 | 0.0292 | No |
| 42 | Soat2 | 9357 | 0.018 | 0.0265 | No |
| 43 | Pex19 | 9569 | 0.010 | 0.0161 | No |
| 44 | Crot | 9714 | 0.004 | 0.0089 | No |
| 45 | Slc27a5 | 9912 | 0.000 | -0.0013 | No |
| 46 | Akr1d1 | 9933 | 0.000 | -0.0023 | No |
| 47 | Acsl5 | 10223 | -0.002 | -0.0171 | No |
| 48 | Npc1 | 10310 | -0.005 | -0.0213 | No |
| 49 | Abca6 | 10504 | -0.010 | -0.0307 | No |
| 50 | Cyp27a1 | 10593 | -0.013 | -0.0346 | No |
| 51 | Abca5 | 10744 | -0.017 | -0.0415 | No |
| 52 | Pex7 | 10752 | -0.018 | -0.0410 | No |
| 53 | Optn | 11408 | -0.044 | -0.0726 | No |
| 54 | Slc29a1 | 11485 | -0.045 | -0.0743 | No |
| 55 | Pex1 | 11572 | -0.048 | -0.0763 | No |
| 56 | Efhc1 | 11756 | -0.056 | -0.0830 | No |
| 57 | Abcd2 | 11794 | -0.057 | -0.0821 | No |
| 58 | Fdxr | 12267 | -0.076 | -0.1027 | No |
| 59 | Slc22a18 | 12341 | -0.078 | -0.1027 | No |
| 60 | Gclm | 12500 | -0.084 | -0.1067 | No |
| 61 | Hsd17b4 | 12826 | -0.097 | -0.1187 | No |
| 62 | Fads2 | 13109 | -0.108 | -0.1280 | No |
| 63 | Atxn1 | 13282 | -0.114 | -0.1313 | No |
| 64 | Pfkm | 13361 | -0.118 | -0.1295 | No |
| 65 | Rbp1 | 14494 | -0.169 | -0.1795 | No |
| 66 | Abca3 | 14504 | -0.170 | -0.1717 | No |
| 67 | Aldh9a1 | 14542 | -0.171 | -0.1652 | No |
| 68 | Idh2 | 14543 | -0.172 | -0.1569 | No |
| 69 | Idh1 | 14667 | -0.177 | -0.1546 | No |
| 70 | Prdx5 | 14898 | -0.190 | -0.1571 | No |
| 71 | Pex11a | 14907 | -0.190 | -0.1483 | No |
| 72 | Sod1 | 15111 | -0.202 | -0.1488 | No |
| 73 | Phyh | 15142 | -0.203 | -0.1404 | No |
| 74 | Isoc1 | 15168 | -0.205 | -0.1317 | No |
| 75 | Retsat | 15197 | -0.206 | -0.1231 | No |
| 76 | Gnpat | 15263 | -0.209 | -0.1162 | No |
| 77 | Cat | 15595 | -0.227 | -0.1222 | No |
| 78 | Hacl1 | 15683 | -0.232 | -0.1153 | No |
| 79 | Aqp9 | 15897 | -0.245 | -0.1143 | No |
| 80 | Pnpla8 | 16401 | -0.272 | -0.1270 | No |
| 81 | Lipe | 16589 | -0.285 | -0.1227 | No |
| 82 | Scp2 | 16704 | -0.293 | -0.1142 | No |
| 83 | Nr1h4 | 17218 | -0.324 | -0.1248 | No |
| 84 | Mlycd | 17321 | -0.329 | -0.1140 | No |
| 85 | Slc35b2 | 17343 | -0.330 | -0.0990 | No |
| 86 | Pecr | 17434 | -0.338 | -0.0871 | No |
| 87 | Hsd3b7 | 18299 | -0.431 | -0.1105 | No |
| 88 | Slc23a1 | 18512 | -0.464 | -0.0988 | No |
| 89 | Pipox | 18708 | -0.496 | -0.0846 | No |
| 90 | Hsd17b11 | 18979 | -0.547 | -0.0718 | No |
| 91 | Gstk1 | 18988 | -0.549 | -0.0454 | No |
| 92 | Bcar3 | 19305 | -0.696 | -0.0276 | No |
| 93 | Paox | 19412 | -0.783 | 0.0051 | No |