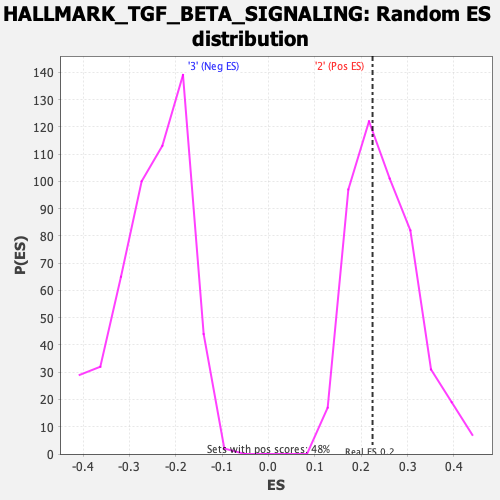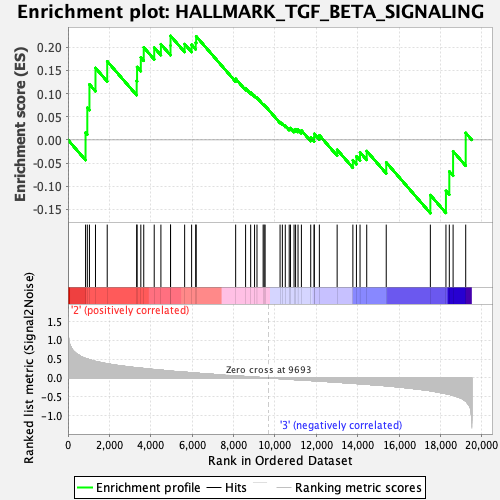
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | 0.22447482 |
| Normalized Enrichment Score (NES) | 0.90711033 |
| Nominal p-value | 0.57983196 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
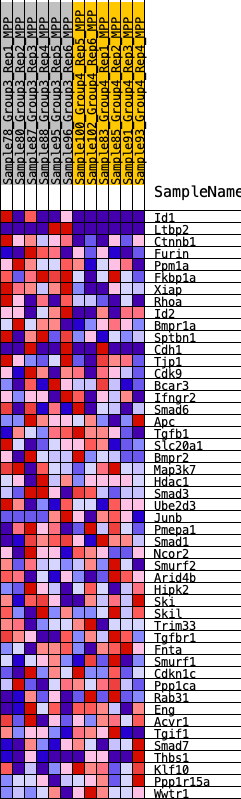
| 1 | Id1 | 848 | 0.512 | 0.0158 | Yes |
| 2 | Ltbp2 | 933 | 0.499 | 0.0695 | Yes |
| 3 | Ctnnb1 | 1036 | 0.479 | 0.1199 | Yes |
| 4 | Furin | 1328 | 0.434 | 0.1553 | Yes |
| 5 | Ppm1a | 1895 | 0.373 | 0.1696 | Yes |
| 6 | Fkbp1a | 3319 | 0.267 | 0.1274 | Yes |
| 7 | Xiap | 3338 | 0.266 | 0.1574 | Yes |
| 8 | Rhoa | 3519 | 0.257 | 0.1779 | Yes |
| 9 | Id2 | 3660 | 0.249 | 0.1996 | Yes |
| 10 | Bmpr1a | 4166 | 0.221 | 0.1993 | Yes |
| 11 | Sptbn1 | 4489 | 0.205 | 0.2065 | Yes |
| 12 | Cdh1 | 4953 | 0.180 | 0.2037 | Yes |
| 13 | Tjp1 | 4956 | 0.180 | 0.2245 | Yes |
| 14 | Cdk9 | 5636 | 0.148 | 0.2068 | No |
| 15 | Bcar3 | 5973 | 0.136 | 0.2054 | No |
| 16 | Ifngr2 | 6171 | 0.127 | 0.2100 | No |
| 17 | Smad6 | 6193 | 0.126 | 0.2235 | No |
| 18 | Apc | 8101 | 0.055 | 0.1319 | No |
| 19 | Tgfb1 | 8581 | 0.037 | 0.1115 | No |
| 20 | Slc20a1 | 8826 | 0.028 | 0.1023 | No |
| 21 | Bmpr2 | 9014 | 0.022 | 0.0952 | No |
| 22 | Map3k7 | 9132 | 0.017 | 0.0912 | No |
| 23 | Hdac1 | 9429 | 0.008 | 0.0769 | No |
| 24 | Smad3 | 9478 | 0.006 | 0.0751 | No |
| 25 | Ube2d3 | 9524 | 0.005 | 0.0735 | No |
| 26 | Junb | 10247 | -0.017 | 0.0383 | No |
| 27 | Pmepa1 | 10358 | -0.020 | 0.0350 | No |
| 28 | Smad1 | 10499 | -0.026 | 0.0308 | No |
| 29 | Ncor2 | 10689 | -0.032 | 0.0247 | No |
| 30 | Smurf2 | 10750 | -0.034 | 0.0257 | No |
| 31 | Arid4b | 10923 | -0.041 | 0.0216 | No |
| 32 | Hipk2 | 10996 | -0.044 | 0.0230 | No |
| 33 | Ski | 11115 | -0.048 | 0.0225 | No |
| 34 | Skil | 11278 | -0.053 | 0.0204 | No |
| 35 | Trim33 | 11728 | -0.067 | 0.0050 | No |
| 36 | Tgfbr1 | 11893 | -0.072 | 0.0050 | No |
| 37 | Fnta | 11899 | -0.073 | 0.0132 | No |
| 38 | Smurf1 | 12146 | -0.081 | 0.0100 | No |
| 39 | Cdkn1c | 13005 | -0.112 | -0.0212 | No |
| 40 | Ppp1ca | 13765 | -0.141 | -0.0438 | No |
| 41 | Rab31 | 13937 | -0.146 | -0.0356 | No |
| 42 | Eng | 14111 | -0.153 | -0.0267 | No |
| 43 | Acvr1 | 14434 | -0.166 | -0.0240 | No |
| 44 | Tgif1 | 15377 | -0.205 | -0.0485 | No |
| 45 | Smad7 | 17509 | -0.336 | -0.1190 | No |
| 46 | Thbs1 | 18262 | -0.412 | -0.1098 | No |
| 47 | Klf10 | 18425 | -0.431 | -0.0681 | No |
| 48 | Ppp1r15a | 18608 | -0.456 | -0.0246 | No |
| 49 | Wwtr1 | 19217 | -0.611 | 0.0152 | No |