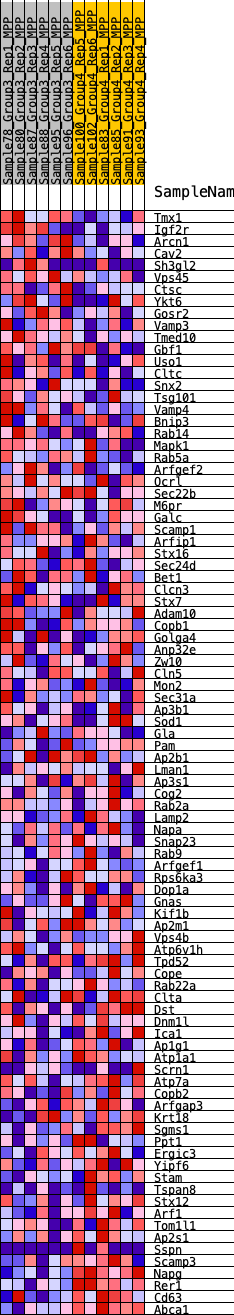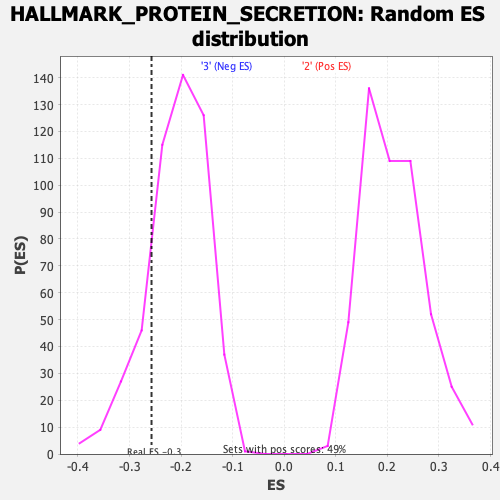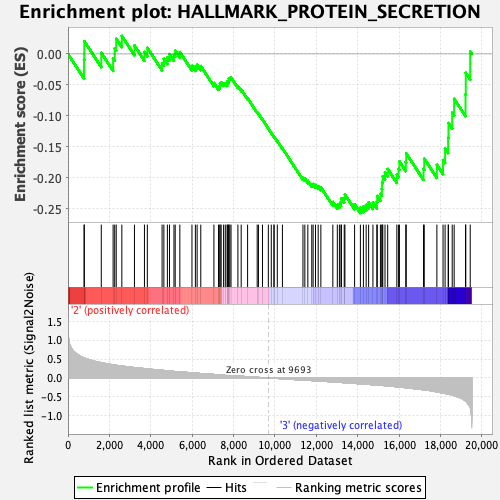
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.2573237 |
| Normalized Enrichment Score (NES) | -1.2405741 |
| Nominal p-value | 0.16798419 |
| FDR q-value | 0.65912896 |
| FWER p-Value | 0.928 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tmx1 | 777 | 0.529 | -0.0092 | No |
| 2 | Igf2r | 788 | 0.525 | 0.0208 | No |
| 3 | Arcn1 | 1608 | 0.403 | 0.0020 | No |
| 4 | Cav2 | 2180 | 0.350 | -0.0071 | No |
| 5 | Sh3gl2 | 2257 | 0.343 | 0.0090 | No |
| 6 | Vps45 | 2330 | 0.335 | 0.0247 | No |
| 7 | Ctsc | 2601 | 0.317 | 0.0292 | No |
| 8 | Ykt6 | 3211 | 0.274 | 0.0138 | No |
| 9 | Gosr2 | 3694 | 0.247 | 0.0033 | No |
| 10 | Vamp3 | 3833 | 0.239 | 0.0101 | No |
| 11 | Tmed10 | 4546 | 0.201 | -0.0149 | No |
| 12 | Gbf1 | 4632 | 0.198 | -0.0078 | No |
| 13 | Uso1 | 4802 | 0.187 | -0.0056 | No |
| 14 | Cltc | 4904 | 0.182 | -0.0002 | No |
| 15 | Snx2 | 5120 | 0.172 | -0.0013 | No |
| 16 | Tsg101 | 5180 | 0.169 | 0.0054 | No |
| 17 | Vamp4 | 5404 | 0.158 | 0.0031 | No |
| 18 | Bnip3 | 5990 | 0.135 | -0.0191 | No |
| 19 | Rab14 | 6155 | 0.128 | -0.0201 | No |
| 20 | Mapk1 | 6239 | 0.124 | -0.0172 | No |
| 21 | Rab5a | 6416 | 0.117 | -0.0195 | No |
| 22 | Arfgef2 | 7051 | 0.092 | -0.0468 | No |
| 23 | Ocrl | 7271 | 0.084 | -0.0532 | No |
| 24 | Sec22b | 7338 | 0.081 | -0.0519 | No |
| 25 | M6pr | 7345 | 0.081 | -0.0475 | No |
| 26 | Galc | 7400 | 0.079 | -0.0457 | No |
| 27 | Scamp1 | 7518 | 0.075 | -0.0474 | No |
| 28 | Arfip1 | 7600 | 0.073 | -0.0473 | No |
| 29 | Stx16 | 7689 | 0.070 | -0.0477 | No |
| 30 | Sec24d | 7702 | 0.070 | -0.0443 | No |
| 31 | Bet1 | 7764 | 0.067 | -0.0436 | No |
| 32 | Clcn3 | 7770 | 0.067 | -0.0399 | No |
| 33 | Stx7 | 7820 | 0.065 | -0.0387 | No |
| 34 | Adam10 | 7877 | 0.063 | -0.0379 | No |
| 35 | Copb1 | 8210 | 0.051 | -0.0521 | No |
| 36 | Golga4 | 8368 | 0.045 | -0.0575 | No |
| 37 | Anp32e | 8678 | 0.033 | -0.0715 | No |
| 38 | Zw10 | 9141 | 0.017 | -0.0943 | No |
| 39 | Cln5 | 9202 | 0.015 | -0.0965 | No |
| 40 | Mon2 | 9397 | 0.009 | -0.1060 | No |
| 41 | Sec31a | 9680 | 0.000 | -0.1205 | No |
| 42 | Ap3b1 | 9822 | -0.002 | -0.1276 | No |
| 43 | Sod1 | 9954 | -0.007 | -0.1340 | No |
| 44 | Gla | 9957 | -0.007 | -0.1337 | No |
| 45 | Pam | 10113 | -0.012 | -0.1410 | No |
| 46 | Ap2b1 | 10361 | -0.020 | -0.1525 | No |
| 47 | Lman1 | 11353 | -0.056 | -0.2003 | No |
| 48 | Ap3s1 | 11444 | -0.059 | -0.2015 | No |
| 49 | Cog2 | 11589 | -0.064 | -0.2052 | No |
| 50 | Rab2a | 11779 | -0.069 | -0.2110 | No |
| 51 | Lamp2 | 11844 | -0.071 | -0.2101 | No |
| 52 | Napa | 11960 | -0.075 | -0.2117 | No |
| 53 | Snap23 | 12088 | -0.079 | -0.2136 | No |
| 54 | Rab9 | 12218 | -0.084 | -0.2154 | No |
| 55 | Arfgef1 | 12797 | -0.104 | -0.2392 | No |
| 56 | Rps6ka3 | 13013 | -0.112 | -0.2437 | No |
| 57 | Dop1a | 13114 | -0.116 | -0.2421 | No |
| 58 | Gnas | 13190 | -0.119 | -0.2391 | No |
| 59 | Kif1b | 13212 | -0.120 | -0.2332 | No |
| 60 | Ap2m1 | 13347 | -0.125 | -0.2329 | No |
| 61 | Vps4b | 13380 | -0.126 | -0.2272 | No |
| 62 | Atp6v1h | 13848 | -0.143 | -0.2430 | No |
| 63 | Tpd52 | 14128 | -0.154 | -0.2484 | Yes |
| 64 | Cope | 14274 | -0.159 | -0.2466 | Yes |
| 65 | Rab22a | 14407 | -0.165 | -0.2438 | Yes |
| 66 | Clta | 14526 | -0.170 | -0.2400 | Yes |
| 67 | Dst | 14734 | -0.179 | -0.2403 | Yes |
| 68 | Dnm1l | 14923 | -0.185 | -0.2392 | Yes |
| 69 | Ica1 | 14943 | -0.185 | -0.2294 | Yes |
| 70 | Ap1g1 | 15096 | -0.191 | -0.2261 | Yes |
| 71 | Atp1a1 | 15164 | -0.195 | -0.2183 | Yes |
| 72 | Scrn1 | 15179 | -0.196 | -0.2076 | Yes |
| 73 | Atp7a | 15210 | -0.197 | -0.1977 | Yes |
| 74 | Copb2 | 15318 | -0.203 | -0.1914 | Yes |
| 75 | Arfgap3 | 15447 | -0.209 | -0.1858 | Yes |
| 76 | Krt18 | 15890 | -0.232 | -0.1951 | Yes |
| 77 | Sgms1 | 15974 | -0.237 | -0.1856 | Yes |
| 78 | Ppt1 | 16014 | -0.239 | -0.1737 | Yes |
| 79 | Ergic3 | 16322 | -0.256 | -0.1746 | Yes |
| 80 | Yipf6 | 16343 | -0.258 | -0.1607 | Yes |
| 81 | Stam | 17177 | -0.310 | -0.1856 | Yes |
| 82 | Tspan8 | 17214 | -0.313 | -0.1692 | Yes |
| 83 | Stx12 | 17824 | -0.369 | -0.1792 | Yes |
| 84 | Arf1 | 18122 | -0.397 | -0.1714 | Yes |
| 85 | Tom1l1 | 18221 | -0.407 | -0.1528 | Yes |
| 86 | Ap2s1 | 18369 | -0.425 | -0.1356 | Yes |
| 87 | Sspn | 18385 | -0.426 | -0.1116 | Yes |
| 88 | Scamp3 | 18565 | -0.450 | -0.0947 | Yes |
| 89 | Napg | 18659 | -0.465 | -0.0725 | Yes |
| 90 | Rer1 | 19208 | -0.606 | -0.0655 | Yes |
| 91 | Cd63 | 19220 | -0.612 | -0.0305 | Yes |
| 92 | Abca1 | 19435 | -0.782 | 0.0040 | Yes |