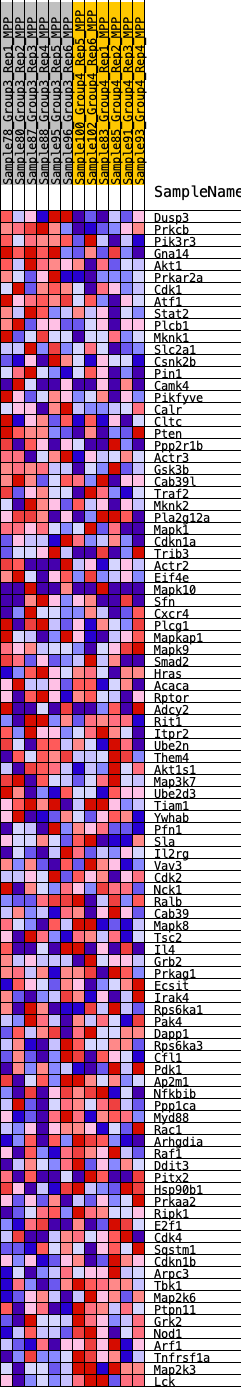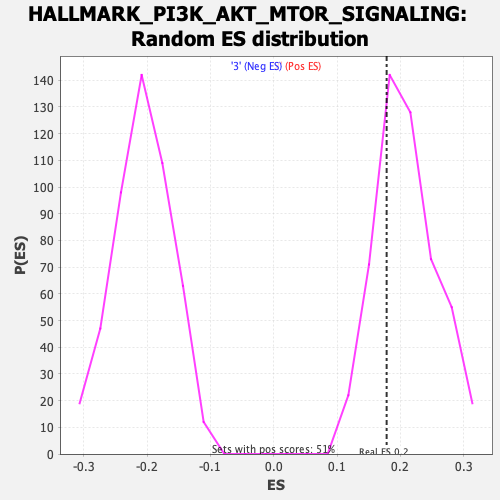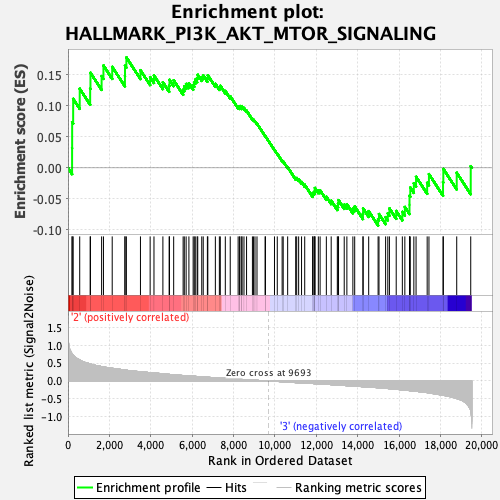
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.17833003 |
| Normalized Enrichment Score (NES) | 0.8536577 |
| Nominal p-value | 0.7352941 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dusp3 | 198 | 0.759 | 0.0319 | Yes |
| 2 | Prkcb | 201 | 0.758 | 0.0739 | Yes |
| 3 | Pik3r3 | 250 | 0.721 | 0.1115 | Yes |
| 4 | Gna14 | 566 | 0.586 | 0.1278 | Yes |
| 5 | Akt1 | 1071 | 0.473 | 0.1281 | Yes |
| 6 | Prkar2a | 1086 | 0.471 | 0.1535 | Yes |
| 7 | Cdk1 | 1624 | 0.401 | 0.1481 | Yes |
| 8 | Atf1 | 1713 | 0.391 | 0.1653 | Yes |
| 9 | Stat2 | 2136 | 0.352 | 0.1631 | Yes |
| 10 | Plcb1 | 2751 | 0.304 | 0.1484 | Yes |
| 11 | Mknk1 | 2755 | 0.304 | 0.1651 | Yes |
| 12 | Slc2a1 | 2821 | 0.298 | 0.1783 | Yes |
| 13 | Csnk2b | 3501 | 0.258 | 0.1577 | No |
| 14 | Pin1 | 3969 | 0.231 | 0.1464 | No |
| 15 | Camk4 | 4154 | 0.221 | 0.1492 | No |
| 16 | Pikfyve | 4585 | 0.200 | 0.1382 | No |
| 17 | Calr | 4887 | 0.183 | 0.1329 | No |
| 18 | Cltc | 4904 | 0.182 | 0.1422 | No |
| 19 | Pten | 5106 | 0.172 | 0.1414 | No |
| 20 | Ppp2r1b | 5563 | 0.152 | 0.1264 | No |
| 21 | Actr3 | 5619 | 0.150 | 0.1319 | No |
| 22 | Gsk3b | 5708 | 0.148 | 0.1356 | No |
| 23 | Cab39l | 5842 | 0.142 | 0.1366 | No |
| 24 | Traf2 | 6047 | 0.133 | 0.1335 | No |
| 25 | Mknk2 | 6100 | 0.130 | 0.1381 | No |
| 26 | Pla2g12a | 6149 | 0.128 | 0.1427 | No |
| 27 | Mapk1 | 6239 | 0.124 | 0.1450 | No |
| 28 | Cdkn1a | 6269 | 0.122 | 0.1503 | No |
| 29 | Trib3 | 6463 | 0.115 | 0.1467 | No |
| 30 | Actr2 | 6534 | 0.112 | 0.1493 | No |
| 31 | Eif4e | 6735 | 0.104 | 0.1448 | No |
| 32 | Mapk10 | 6756 | 0.103 | 0.1495 | No |
| 33 | Sfn | 7120 | 0.090 | 0.1357 | No |
| 34 | Cxcr4 | 7314 | 0.082 | 0.1304 | No |
| 35 | Plcg1 | 7359 | 0.080 | 0.1326 | No |
| 36 | Mapkap1 | 7601 | 0.073 | 0.1242 | No |
| 37 | Mapk9 | 7839 | 0.064 | 0.1155 | No |
| 38 | Smad2 | 8221 | 0.051 | 0.0987 | No |
| 39 | Hras | 8294 | 0.048 | 0.0977 | No |
| 40 | Acaca | 8304 | 0.047 | 0.0998 | No |
| 41 | Rptor | 8400 | 0.043 | 0.0973 | No |
| 42 | Adcy2 | 8403 | 0.043 | 0.0996 | No |
| 43 | Rit1 | 8503 | 0.040 | 0.0967 | No |
| 44 | Itpr2 | 8628 | 0.035 | 0.0923 | No |
| 45 | Ube2n | 8911 | 0.025 | 0.0792 | No |
| 46 | Them4 | 8966 | 0.023 | 0.0777 | No |
| 47 | Akt1s1 | 9035 | 0.021 | 0.0753 | No |
| 48 | Map3k7 | 9132 | 0.017 | 0.0714 | No |
| 49 | Ube2d3 | 9524 | 0.005 | 0.0515 | No |
| 50 | Tiam1 | 9541 | 0.005 | 0.0510 | No |
| 51 | Ywhab | 9977 | -0.007 | 0.0290 | No |
| 52 | Pfn1 | 10112 | -0.012 | 0.0227 | No |
| 53 | Sla | 10347 | -0.020 | 0.0118 | No |
| 54 | Il2rg | 10398 | -0.022 | 0.0104 | No |
| 55 | Vav3 | 10614 | -0.029 | 0.0010 | No |
| 56 | Cdk2 | 11002 | -0.044 | -0.0165 | No |
| 57 | Nck1 | 11050 | -0.046 | -0.0164 | No |
| 58 | Ralb | 11145 | -0.049 | -0.0185 | No |
| 59 | Cab39 | 11285 | -0.054 | -0.0227 | No |
| 60 | Mapk8 | 11438 | -0.059 | -0.0273 | No |
| 61 | Tsc2 | 11818 | -0.070 | -0.0429 | No |
| 62 | Il4 | 11841 | -0.071 | -0.0401 | No |
| 63 | Grb2 | 11900 | -0.073 | -0.0391 | No |
| 64 | Prkag1 | 11927 | -0.073 | -0.0364 | No |
| 65 | Ecsit | 11931 | -0.074 | -0.0324 | No |
| 66 | Irak4 | 12096 | -0.080 | -0.0364 | No |
| 67 | Rps6ka1 | 12182 | -0.082 | -0.0363 | No |
| 68 | Pak4 | 12483 | -0.093 | -0.0466 | No |
| 69 | Dapp1 | 12717 | -0.101 | -0.0530 | No |
| 70 | Rps6ka3 | 13013 | -0.112 | -0.0619 | No |
| 71 | Cfl1 | 13049 | -0.114 | -0.0574 | No |
| 72 | Pdk1 | 13072 | -0.114 | -0.0522 | No |
| 73 | Ap2m1 | 13347 | -0.125 | -0.0594 | No |
| 74 | Nfkbib | 13475 | -0.130 | -0.0587 | No |
| 75 | Ppp1ca | 13765 | -0.141 | -0.0658 | No |
| 76 | Myd88 | 13853 | -0.143 | -0.0623 | No |
| 77 | Rac1 | 14252 | -0.159 | -0.0740 | No |
| 78 | Arhgdia | 14260 | -0.159 | -0.0655 | No |
| 79 | Raf1 | 14529 | -0.170 | -0.0699 | No |
| 80 | Ddit3 | 14984 | -0.187 | -0.0829 | No |
| 81 | Pitx2 | 15027 | -0.189 | -0.0745 | No |
| 82 | Hsp90b1 | 15347 | -0.204 | -0.0796 | No |
| 83 | Prkaa2 | 15453 | -0.210 | -0.0734 | No |
| 84 | Ripk1 | 15531 | -0.214 | -0.0655 | No |
| 85 | E2f1 | 15859 | -0.230 | -0.0696 | No |
| 86 | Cdk4 | 16155 | -0.248 | -0.0710 | No |
| 87 | Sqstm1 | 16276 | -0.254 | -0.0631 | No |
| 88 | Cdkn1b | 16501 | -0.268 | -0.0597 | No |
| 89 | Arpc3 | 16502 | -0.268 | -0.0448 | No |
| 90 | Tbk1 | 16539 | -0.271 | -0.0317 | No |
| 91 | Map2k6 | 16711 | -0.281 | -0.0249 | No |
| 92 | Ptpn11 | 16818 | -0.288 | -0.0144 | No |
| 93 | Grk2 | 17355 | -0.324 | -0.0240 | No |
| 94 | Nod1 | 17441 | -0.329 | -0.0101 | No |
| 95 | Arf1 | 18122 | -0.397 | -0.0231 | No |
| 96 | Tnfrsf1a | 18136 | -0.398 | -0.0017 | No |
| 97 | Map2k3 | 18783 | -0.487 | -0.0079 | No |
| 98 | Lck | 19460 | -0.817 | 0.0027 | No |