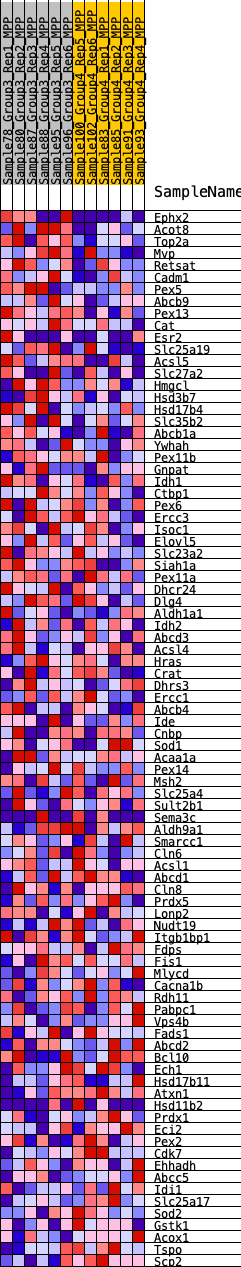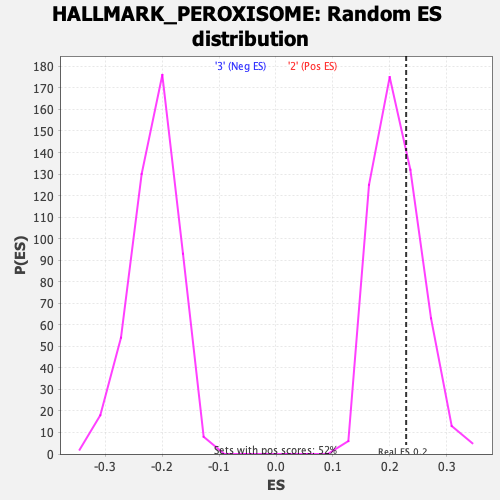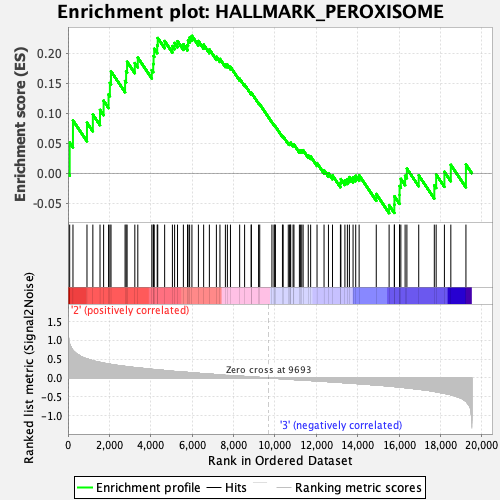
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.22871435 |
| Normalized Enrichment Score (NES) | 1.0749774 |
| Nominal p-value | 0.34874758 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ephx2 | 81 | 0.889 | 0.0510 | Yes |
| 2 | Acot8 | 240 | 0.725 | 0.0879 | Yes |
| 3 | Top2a | 917 | 0.502 | 0.0842 | Yes |
| 4 | Mvp | 1201 | 0.453 | 0.0978 | Yes |
| 5 | Retsat | 1547 | 0.410 | 0.1055 | Yes |
| 6 | Cadm1 | 1721 | 0.390 | 0.1207 | Yes |
| 7 | Pex5 | 1960 | 0.367 | 0.1312 | Yes |
| 8 | Abcb9 | 2024 | 0.362 | 0.1505 | Yes |
| 9 | Pex13 | 2078 | 0.356 | 0.1698 | Yes |
| 10 | Cat | 2758 | 0.304 | 0.1537 | Yes |
| 11 | Esr2 | 2808 | 0.299 | 0.1698 | Yes |
| 12 | Slc25a19 | 2855 | 0.296 | 0.1857 | Yes |
| 13 | Acsl5 | 3229 | 0.272 | 0.1834 | Yes |
| 14 | Slc27a2 | 3373 | 0.264 | 0.1924 | Yes |
| 15 | Hmgcl | 4050 | 0.226 | 0.1716 | Yes |
| 16 | Hsd3b7 | 4118 | 0.224 | 0.1821 | Yes |
| 17 | Hsd17b4 | 4135 | 0.223 | 0.1951 | Yes |
| 18 | Slc35b2 | 4165 | 0.221 | 0.2073 | Yes |
| 19 | Abcb1a | 4311 | 0.215 | 0.2132 | Yes |
| 20 | Ywhah | 4336 | 0.213 | 0.2252 | Yes |
| 21 | Pex11b | 4670 | 0.196 | 0.2201 | Yes |
| 22 | Gnpat | 5045 | 0.175 | 0.2118 | Yes |
| 23 | Idh1 | 5151 | 0.170 | 0.2169 | Yes |
| 24 | Ctbp1 | 5292 | 0.163 | 0.2198 | Yes |
| 25 | Pex6 | 5575 | 0.152 | 0.2147 | Yes |
| 26 | Ercc3 | 5774 | 0.145 | 0.2135 | Yes |
| 27 | Isoc1 | 5800 | 0.144 | 0.2212 | Yes |
| 28 | Elovl5 | 5876 | 0.141 | 0.2260 | Yes |
| 29 | Slc23a2 | 5988 | 0.135 | 0.2287 | Yes |
| 30 | Siah1a | 6301 | 0.121 | 0.2202 | No |
| 31 | Pex11a | 6554 | 0.111 | 0.2141 | No |
| 32 | Dhcr24 | 6831 | 0.100 | 0.2061 | No |
| 33 | Dlg4 | 7170 | 0.087 | 0.1941 | No |
| 34 | Aldh1a1 | 7342 | 0.081 | 0.1903 | No |
| 35 | Idh2 | 7605 | 0.073 | 0.1813 | No |
| 36 | Abcd3 | 7701 | 0.070 | 0.1808 | No |
| 37 | Acsl4 | 7850 | 0.064 | 0.1771 | No |
| 38 | Hras | 8294 | 0.048 | 0.1572 | No |
| 39 | Crat | 8533 | 0.038 | 0.1474 | No |
| 40 | Dhrs3 | 8852 | 0.027 | 0.1327 | No |
| 41 | Ercc1 | 8859 | 0.027 | 0.1340 | No |
| 42 | Abcb4 | 9209 | 0.015 | 0.1170 | No |
| 43 | Ide | 9264 | 0.013 | 0.1150 | No |
| 44 | Cnbp | 9854 | -0.003 | 0.0849 | No |
| 45 | Sod1 | 9954 | -0.007 | 0.0802 | No |
| 46 | Acaa1a | 9987 | -0.008 | 0.0790 | No |
| 47 | Pex14 | 10022 | -0.008 | 0.0778 | No |
| 48 | Msh2 | 10362 | -0.020 | 0.0616 | No |
| 49 | Slc25a4 | 10396 | -0.022 | 0.0613 | No |
| 50 | Sult2b1 | 10644 | -0.030 | 0.0504 | No |
| 51 | Sema3c | 10709 | -0.033 | 0.0491 | No |
| 52 | Aldh9a1 | 10735 | -0.034 | 0.0499 | No |
| 53 | Smarcc1 | 10761 | -0.035 | 0.0508 | No |
| 54 | Cln6 | 10875 | -0.039 | 0.0474 | No |
| 55 | Acsl1 | 10918 | -0.041 | 0.0478 | No |
| 56 | Abcd1 | 11181 | -0.050 | 0.0374 | No |
| 57 | Cln8 | 11240 | -0.052 | 0.0376 | No |
| 58 | Prdx5 | 11296 | -0.054 | 0.0382 | No |
| 59 | Lonp2 | 11361 | -0.056 | 0.0384 | No |
| 60 | Nudt19 | 11609 | -0.065 | 0.0296 | No |
| 61 | Itgb1bp1 | 11724 | -0.067 | 0.0279 | No |
| 62 | Fdps | 12033 | -0.077 | 0.0169 | No |
| 63 | Fis1 | 12372 | -0.089 | 0.0050 | No |
| 64 | Mlycd | 12585 | -0.096 | 0.0000 | No |
| 65 | Cacna1b | 12781 | -0.103 | -0.0036 | No |
| 66 | Rdh11 | 13163 | -0.118 | -0.0159 | No |
| 67 | Pabpc1 | 13186 | -0.119 | -0.0097 | No |
| 68 | Vps4b | 13380 | -0.126 | -0.0118 | No |
| 69 | Fads1 | 13500 | -0.131 | -0.0098 | No |
| 70 | Abcd2 | 13597 | -0.135 | -0.0064 | No |
| 71 | Bcl10 | 13774 | -0.141 | -0.0067 | No |
| 72 | Ech1 | 13902 | -0.145 | -0.0043 | No |
| 73 | Hsd17b11 | 14067 | -0.151 | -0.0033 | No |
| 74 | Atxn1 | 14895 | -0.183 | -0.0345 | No |
| 75 | Hsd11b2 | 15516 | -0.213 | -0.0532 | No |
| 76 | Prdx1 | 15767 | -0.225 | -0.0521 | No |
| 77 | Eci2 | 15774 | -0.226 | -0.0384 | No |
| 78 | Pex2 | 16019 | -0.239 | -0.0361 | No |
| 79 | Cdk7 | 16022 | -0.240 | -0.0214 | No |
| 80 | Ehhadh | 16083 | -0.243 | -0.0094 | No |
| 81 | Abcc5 | 16289 | -0.255 | -0.0041 | No |
| 82 | Idi1 | 16375 | -0.260 | 0.0076 | No |
| 83 | Slc25a17 | 16947 | -0.295 | -0.0034 | No |
| 84 | Sod2 | 17700 | -0.354 | -0.0202 | No |
| 85 | Gstk1 | 17789 | -0.364 | -0.0021 | No |
| 86 | Acox1 | 18188 | -0.404 | 0.0025 | No |
| 87 | Tspo | 18498 | -0.442 | 0.0140 | No |
| 88 | Scp2 | 19228 | -0.615 | 0.0146 | No |