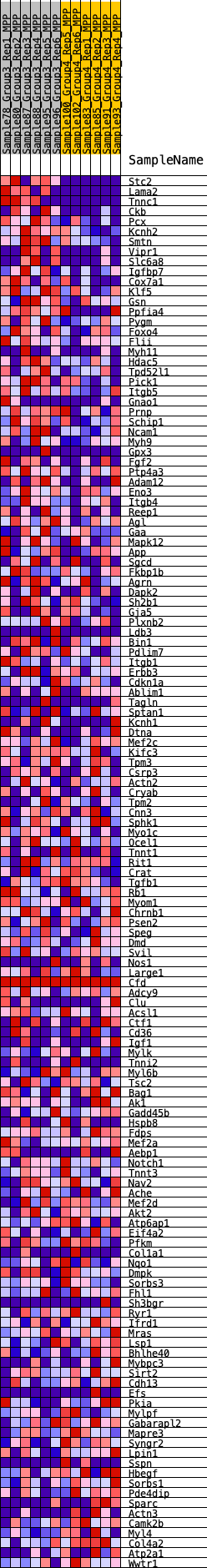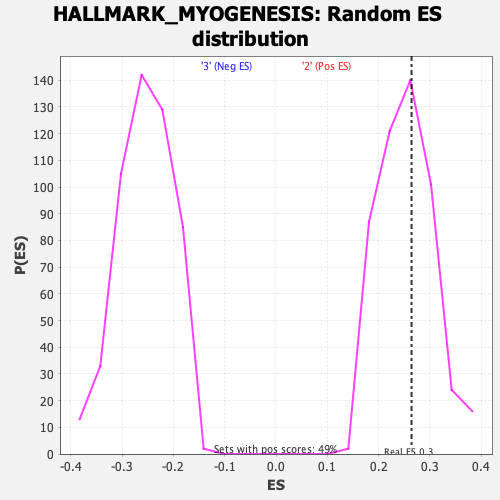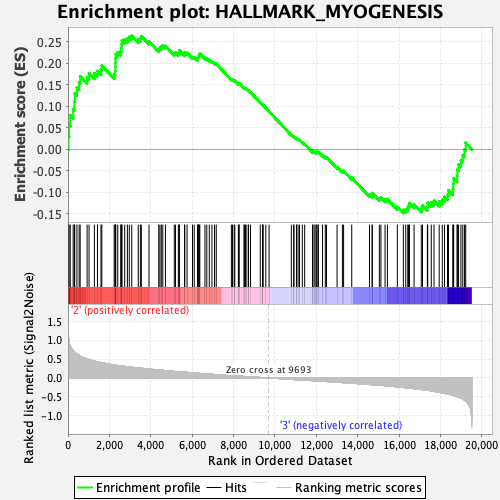
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.26413882 |
| Normalized Enrichment Score (NES) | 1.0391887 |
| Nominal p-value | 0.40733197 |
| FDR q-value | 0.9410949 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Stc2 | 25 | 1.071 | 0.0305 | Yes |
| 2 | Lama2 | 53 | 0.938 | 0.0569 | Yes |
| 3 | Tnnc1 | 108 | 0.858 | 0.0796 | Yes |
| 4 | Ckb | 249 | 0.721 | 0.0937 | Yes |
| 5 | Pcx | 309 | 0.686 | 0.1110 | Yes |
| 6 | Kcnh2 | 325 | 0.679 | 0.1304 | Yes |
| 7 | Smtn | 431 | 0.627 | 0.1436 | Yes |
| 8 | Vipr1 | 537 | 0.595 | 0.1558 | Yes |
| 9 | Slc6a8 | 587 | 0.578 | 0.1704 | Yes |
| 10 | Igfbp7 | 923 | 0.501 | 0.1680 | Yes |
| 11 | Cox7a1 | 1017 | 0.482 | 0.1775 | Yes |
| 12 | Klf5 | 1274 | 0.441 | 0.1773 | Yes |
| 13 | Gsn | 1424 | 0.424 | 0.1822 | Yes |
| 14 | Ppfia4 | 1594 | 0.404 | 0.1855 | Yes |
| 15 | Pygm | 1633 | 0.399 | 0.1954 | Yes |
| 16 | Foxo4 | 2231 | 0.346 | 0.1748 | Yes |
| 17 | Flii | 2275 | 0.341 | 0.1827 | Yes |
| 18 | Myh11 | 2289 | 0.340 | 0.1921 | Yes |
| 19 | Hdac5 | 2292 | 0.339 | 0.2021 | Yes |
| 20 | Tpd52l1 | 2294 | 0.339 | 0.2121 | Yes |
| 21 | Pick1 | 2309 | 0.337 | 0.2214 | Yes |
| 22 | Itgb5 | 2404 | 0.328 | 0.2263 | Yes |
| 23 | Gnao1 | 2548 | 0.321 | 0.2284 | Yes |
| 24 | Prnp | 2578 | 0.318 | 0.2363 | Yes |
| 25 | Schip1 | 2595 | 0.317 | 0.2449 | Yes |
| 26 | Ncam1 | 2606 | 0.316 | 0.2538 | Yes |
| 27 | Myh9 | 2740 | 0.305 | 0.2559 | Yes |
| 28 | Gpx3 | 2870 | 0.294 | 0.2580 | Yes |
| 29 | Fgf2 | 2961 | 0.289 | 0.2620 | Yes |
| 30 | Ptp4a3 | 3081 | 0.281 | 0.2641 | Yes |
| 31 | Adam12 | 3394 | 0.262 | 0.2558 | No |
| 32 | Eno3 | 3497 | 0.258 | 0.2582 | No |
| 33 | Itgb4 | 3543 | 0.256 | 0.2635 | No |
| 34 | Reep1 | 3914 | 0.234 | 0.2513 | No |
| 35 | Agl | 4384 | 0.211 | 0.2333 | No |
| 36 | Gaa | 4442 | 0.208 | 0.2366 | No |
| 37 | Mapk12 | 4507 | 0.204 | 0.2393 | No |
| 38 | App | 4580 | 0.201 | 0.2415 | No |
| 39 | Sgcd | 4708 | 0.193 | 0.2407 | No |
| 40 | Fkbp1b | 5134 | 0.171 | 0.2238 | No |
| 41 | Agrn | 5195 | 0.168 | 0.2257 | No |
| 42 | Dapk2 | 5332 | 0.161 | 0.2235 | No |
| 43 | Sh2b1 | 5373 | 0.159 | 0.2261 | No |
| 44 | Gja5 | 5378 | 0.159 | 0.2306 | No |
| 45 | Plxnb2 | 5631 | 0.149 | 0.2220 | No |
| 46 | Ldb3 | 5643 | 0.148 | 0.2259 | No |
| 47 | Bin1 | 5753 | 0.146 | 0.2246 | No |
| 48 | Pdlim7 | 6015 | 0.134 | 0.2151 | No |
| 49 | Itgb1 | 6099 | 0.131 | 0.2147 | No |
| 50 | Erbb3 | 6256 | 0.123 | 0.2103 | No |
| 51 | Cdkn1a | 6269 | 0.122 | 0.2133 | No |
| 52 | Ablim1 | 6294 | 0.121 | 0.2156 | No |
| 53 | Tagln | 6324 | 0.120 | 0.2177 | No |
| 54 | Sptan1 | 6340 | 0.120 | 0.2205 | No |
| 55 | Kcnh1 | 6369 | 0.119 | 0.2226 | No |
| 56 | Dtna | 6618 | 0.109 | 0.2130 | No |
| 57 | Mef2c | 6707 | 0.105 | 0.2115 | No |
| 58 | Kifc3 | 6827 | 0.100 | 0.2084 | No |
| 59 | Tpm3 | 6956 | 0.096 | 0.2046 | No |
| 60 | Csrp3 | 7081 | 0.091 | 0.2009 | No |
| 61 | Actn2 | 7163 | 0.088 | 0.1993 | No |
| 62 | Cryab | 7894 | 0.062 | 0.1635 | No |
| 63 | Tpm2 | 7947 | 0.060 | 0.1626 | No |
| 64 | Cnn3 | 8029 | 0.058 | 0.1601 | No |
| 65 | Sphk1 | 8060 | 0.056 | 0.1602 | No |
| 66 | Myo1c | 8231 | 0.050 | 0.1530 | No |
| 67 | Ocel1 | 8260 | 0.049 | 0.1530 | No |
| 68 | Tnnt1 | 8266 | 0.049 | 0.1542 | No |
| 69 | Rit1 | 8503 | 0.040 | 0.1431 | No |
| 70 | Crat | 8533 | 0.038 | 0.1428 | No |
| 71 | Tgfb1 | 8581 | 0.037 | 0.1415 | No |
| 72 | Rb1 | 8604 | 0.036 | 0.1414 | No |
| 73 | Myom1 | 8711 | 0.032 | 0.1369 | No |
| 74 | Chrnb1 | 8717 | 0.032 | 0.1376 | No |
| 75 | Psen2 | 8819 | 0.028 | 0.1332 | No |
| 76 | Speg | 9291 | 0.012 | 0.1092 | No |
| 77 | Dmd | 9410 | 0.008 | 0.1034 | No |
| 78 | Svil | 9415 | 0.008 | 0.1034 | No |
| 79 | Nos1 | 9417 | 0.008 | 0.1036 | No |
| 80 | Large1 | 9557 | 0.004 | 0.0965 | No |
| 81 | Cfd | 9716 | 0.000 | 0.0884 | No |
| 82 | Adcy9 | 10791 | -0.036 | 0.0340 | No |
| 83 | Clu | 10907 | -0.041 | 0.0293 | No |
| 84 | Acsl1 | 10918 | -0.041 | 0.0300 | No |
| 85 | Ctf1 | 11040 | -0.045 | 0.0251 | No |
| 86 | Cd36 | 11045 | -0.045 | 0.0262 | No |
| 87 | Igf1 | 11118 | -0.048 | 0.0239 | No |
| 88 | Mylk | 11185 | -0.050 | 0.0220 | No |
| 89 | Tnni2 | 11322 | -0.055 | 0.0166 | No |
| 90 | Myl6b | 11436 | -0.059 | 0.0125 | No |
| 91 | Tsc2 | 11818 | -0.070 | -0.0051 | No |
| 92 | Bag1 | 11828 | -0.070 | -0.0035 | No |
| 93 | Ak1 | 11895 | -0.072 | -0.0047 | No |
| 94 | Gadd45b | 11976 | -0.075 | -0.0066 | No |
| 95 | Hspb8 | 12022 | -0.077 | -0.0066 | No |
| 96 | Fdps | 12033 | -0.077 | -0.0049 | No |
| 97 | Mef2a | 12100 | -0.080 | -0.0059 | No |
| 98 | Aebp1 | 12301 | -0.086 | -0.0137 | No |
| 99 | Notch1 | 12433 | -0.091 | -0.0177 | No |
| 100 | Tnnt3 | 12491 | -0.093 | -0.0179 | No |
| 101 | Nav2 | 13001 | -0.111 | -0.0409 | No |
| 102 | Ache | 13259 | -0.121 | -0.0506 | No |
| 103 | Mef2d | 13317 | -0.123 | -0.0499 | No |
| 104 | Akt2 | 13710 | -0.139 | -0.0660 | No |
| 105 | Atp6ap1 | 14568 | -0.172 | -0.1051 | No |
| 106 | Eif4a2 | 14697 | -0.177 | -0.1064 | No |
| 107 | Pfkm | 14702 | -0.178 | -0.1014 | No |
| 108 | Col1a1 | 15044 | -0.189 | -0.1134 | No |
| 109 | Nqo1 | 15119 | -0.192 | -0.1115 | No |
| 110 | Dmpk | 15320 | -0.203 | -0.1158 | No |
| 111 | Sorbs3 | 15433 | -0.209 | -0.1154 | No |
| 112 | Fhl1 | 15913 | -0.233 | -0.1332 | No |
| 113 | Sh3bgr | 16204 | -0.250 | -0.1407 | No |
| 114 | Ryr1 | 16320 | -0.256 | -0.1391 | No |
| 115 | Ifrd1 | 16418 | -0.262 | -0.1363 | No |
| 116 | Mras | 16457 | -0.264 | -0.1304 | No |
| 117 | Lsp1 | 16499 | -0.268 | -0.1246 | No |
| 118 | Bhlhe40 | 16722 | -0.281 | -0.1277 | No |
| 119 | Mybpc3 | 17070 | -0.304 | -0.1366 | No |
| 120 | Sirt2 | 17129 | -0.308 | -0.1305 | No |
| 121 | Cdh13 | 17364 | -0.325 | -0.1329 | No |
| 122 | Efs | 17382 | -0.326 | -0.1241 | No |
| 123 | Pkia | 17553 | -0.342 | -0.1228 | No |
| 124 | Mylpf | 17686 | -0.353 | -0.1191 | No |
| 125 | Gabarapl2 | 17935 | -0.379 | -0.1207 | No |
| 126 | Mapre3 | 18085 | -0.393 | -0.1167 | No |
| 127 | Syngr2 | 18192 | -0.404 | -0.1102 | No |
| 128 | Lpin1 | 18337 | -0.421 | -0.1051 | No |
| 129 | Sspn | 18385 | -0.426 | -0.0949 | No |
| 130 | Hbegf | 18595 | -0.454 | -0.0923 | No |
| 131 | Sorbs1 | 18615 | -0.457 | -0.0797 | No |
| 132 | Pde4dip | 18636 | -0.461 | -0.0670 | No |
| 133 | Sparc | 18802 | -0.493 | -0.0609 | No |
| 134 | Actn3 | 18812 | -0.494 | -0.0468 | No |
| 135 | Camk2b | 18872 | -0.505 | -0.0348 | No |
| 136 | Myl4 | 18987 | -0.527 | -0.0251 | No |
| 137 | Col4a2 | 19077 | -0.556 | -0.0132 | No |
| 138 | Atp2a1 | 19161 | -0.587 | -0.0001 | No |
| 139 | Wwtr1 | 19217 | -0.611 | 0.0152 | No |