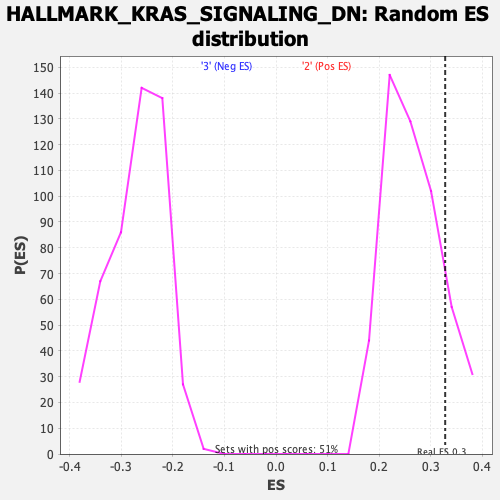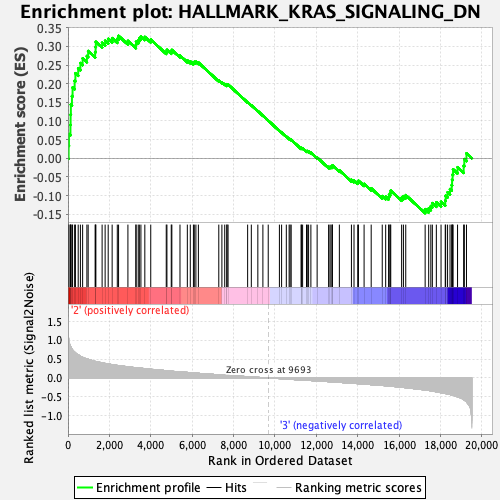
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.3281443 |
| Normalized Enrichment Score (NES) | 1.2327732 |
| Nominal p-value | 0.1509804 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.928 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Epha5 | 31 | 1.048 | 0.0335 | Yes |
| 2 | Snn | 46 | 0.960 | 0.0649 | Yes |
| 3 | Tent5c | 118 | 0.842 | 0.0895 | Yes |
| 4 | Itgb1bp2 | 126 | 0.831 | 0.1170 | Yes |
| 5 | Tfcp2l1 | 138 | 0.820 | 0.1439 | Yes |
| 6 | Tg | 191 | 0.762 | 0.1667 | Yes |
| 7 | Abcg4 | 221 | 0.736 | 0.1899 | Yes |
| 8 | Gp1ba | 321 | 0.681 | 0.2076 | Yes |
| 9 | Sptbn2 | 358 | 0.662 | 0.2279 | Yes |
| 10 | Camk1d | 493 | 0.608 | 0.2414 | Yes |
| 11 | Capn9 | 599 | 0.574 | 0.2552 | Yes |
| 12 | Celsr2 | 707 | 0.543 | 0.2679 | Yes |
| 13 | Brdt | 913 | 0.503 | 0.2741 | Yes |
| 14 | Fgfr3 | 974 | 0.492 | 0.2875 | Yes |
| 15 | Bard1 | 1311 | 0.436 | 0.2848 | Yes |
| 16 | Arpp21 | 1333 | 0.433 | 0.2982 | Yes |
| 17 | Kcnn1 | 1344 | 0.432 | 0.3122 | Yes |
| 18 | Vps50 | 1648 | 0.397 | 0.3099 | Yes |
| 19 | Pde6b | 1794 | 0.383 | 0.3152 | Yes |
| 20 | Zfp112 | 1942 | 0.369 | 0.3200 | Yes |
| 21 | Col2a1 | 2133 | 0.352 | 0.3220 | Yes |
| 22 | Macroh2a2 | 2382 | 0.330 | 0.3203 | Yes |
| 23 | Cacna1f | 2443 | 0.326 | 0.3281 | Yes |
| 24 | Plag1 | 2895 | 0.293 | 0.3147 | No |
| 25 | Cd80 | 3280 | 0.268 | 0.3039 | No |
| 26 | Htr1b | 3289 | 0.268 | 0.3125 | No |
| 27 | Lfng | 3393 | 0.262 | 0.3160 | No |
| 28 | Arhgdig | 3447 | 0.260 | 0.3219 | No |
| 29 | Tcf7l1 | 3523 | 0.257 | 0.3267 | No |
| 30 | Mthfr | 3712 | 0.246 | 0.3252 | No |
| 31 | Gtf3c5 | 3998 | 0.229 | 0.3182 | No |
| 32 | Tgfb2 | 4747 | 0.191 | 0.2861 | No |
| 33 | P2rx6 | 4780 | 0.188 | 0.2907 | No |
| 34 | Slc6a3 | 4989 | 0.178 | 0.2860 | No |
| 35 | Zc2hc1c | 5018 | 0.177 | 0.2904 | No |
| 36 | Efhd1 | 5411 | 0.158 | 0.2755 | No |
| 37 | Idua | 5766 | 0.145 | 0.2621 | No |
| 38 | Cpeb3 | 5909 | 0.139 | 0.2595 | No |
| 39 | Nrip2 | 6060 | 0.133 | 0.2562 | No |
| 40 | Htr1d | 6102 | 0.130 | 0.2584 | No |
| 41 | Pdk2 | 6180 | 0.126 | 0.2587 | No |
| 42 | Msh5 | 6300 | 0.121 | 0.2566 | No |
| 43 | Clstn3 | 7286 | 0.083 | 0.2087 | No |
| 44 | Slc16a7 | 7436 | 0.078 | 0.2036 | No |
| 45 | Lgals7 | 7573 | 0.074 | 0.1991 | No |
| 46 | Ypel1 | 7676 | 0.071 | 0.1962 | No |
| 47 | Thrb | 7679 | 0.070 | 0.1984 | No |
| 48 | Fggy | 7734 | 0.068 | 0.1979 | No |
| 49 | Copz2 | 8680 | 0.033 | 0.1504 | No |
| 50 | Ccdc106 | 8858 | 0.027 | 0.1421 | No |
| 51 | Myo15a | 9173 | 0.016 | 0.1265 | No |
| 52 | Nos1 | 9417 | 0.008 | 0.1142 | No |
| 53 | Btg2 | 9675 | 0.001 | 0.1010 | No |
| 54 | Slc25a23 | 10215 | -0.016 | 0.0738 | No |
| 55 | Ptprj | 10321 | -0.018 | 0.0690 | No |
| 56 | Selenop | 10549 | -0.027 | 0.0582 | No |
| 57 | Zbtb16 | 10685 | -0.032 | 0.0523 | No |
| 58 | Ngb | 10732 | -0.034 | 0.0511 | No |
| 59 | Thnsl2 | 10784 | -0.036 | 0.0496 | No |
| 60 | Chst2 | 11264 | -0.053 | 0.0267 | No |
| 61 | Skil | 11278 | -0.053 | 0.0278 | No |
| 62 | Nr6a1 | 11336 | -0.055 | 0.0267 | No |
| 63 | Mefv | 11524 | -0.062 | 0.0192 | No |
| 64 | Kmt2d | 11561 | -0.063 | 0.0194 | No |
| 65 | Stag3 | 11625 | -0.065 | 0.0184 | No |
| 66 | Tnni3 | 11736 | -0.067 | 0.0149 | No |
| 67 | Edar | 12038 | -0.078 | 0.0020 | No |
| 68 | Mast3 | 12592 | -0.096 | -0.0232 | No |
| 69 | Dcc | 12657 | -0.098 | -0.0232 | No |
| 70 | Gpr19 | 12713 | -0.101 | -0.0227 | No |
| 71 | Cyp39a1 | 12773 | -0.103 | -0.0223 | No |
| 72 | Prodh | 12775 | -0.103 | -0.0189 | No |
| 73 | Tgm1 | 13112 | -0.116 | -0.0323 | No |
| 74 | Synpo | 13695 | -0.138 | -0.0577 | No |
| 75 | Slc29a3 | 13820 | -0.142 | -0.0593 | No |
| 76 | Egf | 14000 | -0.149 | -0.0636 | No |
| 77 | Coq8a | 14034 | -0.150 | -0.0602 | No |
| 78 | Cdkal1 | 14307 | -0.161 | -0.0689 | No |
| 79 | Sphk2 | 14650 | -0.175 | -0.0806 | No |
| 80 | Ryr2 | 15184 | -0.196 | -0.1015 | No |
| 81 | Klk8 | 15350 | -0.204 | -0.1032 | No |
| 82 | Asb7 | 15492 | -0.212 | -0.1033 | No |
| 83 | Hsd11b2 | 15516 | -0.213 | -0.0974 | No |
| 84 | Prkn | 15569 | -0.216 | -0.0929 | No |
| 85 | Dtnb | 15595 | -0.217 | -0.0869 | No |
| 86 | Atp4a | 16121 | -0.245 | -0.1057 | No |
| 87 | Ntf3 | 16207 | -0.250 | -0.1017 | No |
| 88 | Ryr1 | 16320 | -0.256 | -0.0989 | No |
| 89 | Nr4a2 | 17257 | -0.316 | -0.1366 | No |
| 90 | Sgk1 | 17428 | -0.328 | -0.1343 | No |
| 91 | Tenm2 | 17532 | -0.339 | -0.1283 | No |
| 92 | Cpa2 | 17605 | -0.347 | -0.1204 | No |
| 93 | Ybx2 | 17798 | -0.365 | -0.1181 | No |
| 94 | Entpd7 | 18025 | -0.389 | -0.1167 | No |
| 95 | Gamt | 18228 | -0.408 | -0.1134 | No |
| 96 | Mx2 | 18248 | -0.411 | -0.1006 | No |
| 97 | Bmpr1b | 18341 | -0.421 | -0.0912 | No |
| 98 | Tex15 | 18459 | -0.436 | -0.0826 | No |
| 99 | Rsad2 | 18542 | -0.447 | -0.0719 | No |
| 100 | Sidt1 | 18559 | -0.449 | -0.0577 | No |
| 101 | Serpinb2 | 18583 | -0.452 | -0.0437 | No |
| 102 | Magix | 18606 | -0.456 | -0.0296 | No |
| 103 | Mfsd6 | 18823 | -0.496 | -0.0241 | No |
| 104 | Rgs11 | 19118 | -0.570 | -0.0202 | No |
| 105 | Kcnd1 | 19150 | -0.580 | -0.0023 | No |
| 106 | Grid2 | 19254 | -0.625 | 0.0133 | No |