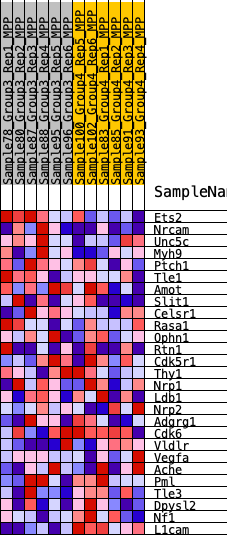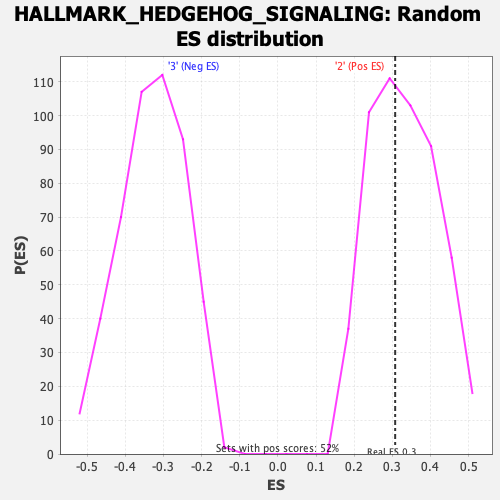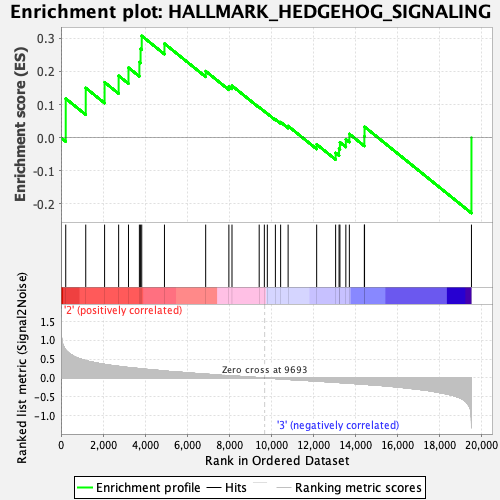
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.30749133 |
| Normalized Enrichment Score (NES) | 0.93176764 |
| Nominal p-value | 0.5606936 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ets2 | 225 | 0.733 | 0.1181 | Yes |
| 2 | Nrcam | 1176 | 0.457 | 0.1503 | Yes |
| 3 | Unc5c | 2072 | 0.357 | 0.1674 | Yes |
| 4 | Myh9 | 2740 | 0.305 | 0.1871 | Yes |
| 5 | Ptch1 | 3208 | 0.274 | 0.2116 | Yes |
| 6 | Tle1 | 3726 | 0.245 | 0.2284 | Yes |
| 7 | Amot | 3783 | 0.241 | 0.2681 | Yes |
| 8 | Slit1 | 3838 | 0.238 | 0.3075 | Yes |
| 9 | Celsr1 | 4918 | 0.181 | 0.2842 | No |
| 10 | Rasa1 | 6877 | 0.098 | 0.2010 | No |
| 11 | Ophn1 | 7980 | 0.059 | 0.1550 | No |
| 12 | Rtn1 | 8128 | 0.054 | 0.1569 | No |
| 13 | Cdk5r1 | 9421 | 0.008 | 0.0920 | No |
| 14 | Thy1 | 9666 | 0.001 | 0.0796 | No |
| 15 | Nrp1 | 9808 | -0.002 | 0.0727 | No |
| 16 | Ldb1 | 10191 | -0.015 | 0.0557 | No |
| 17 | Nrp2 | 10440 | -0.023 | 0.0471 | No |
| 18 | Adgrg1 | 10796 | -0.036 | 0.0353 | No |
| 19 | Cdk6 | 12153 | -0.081 | -0.0199 | No |
| 20 | Vldlr | 13054 | -0.114 | -0.0460 | No |
| 21 | Vegfa | 13218 | -0.120 | -0.0332 | No |
| 22 | Ache | 13259 | -0.121 | -0.0138 | No |
| 23 | Pml | 13540 | -0.133 | -0.0047 | No |
| 24 | Tle3 | 13707 | -0.139 | 0.0114 | No |
| 25 | Dpysl2 | 14419 | -0.165 | 0.0041 | No |
| 26 | Nf1 | 14429 | -0.166 | 0.0329 | No |
| 27 | L1cam | 19511 | -1.289 | 0.0001 | No |