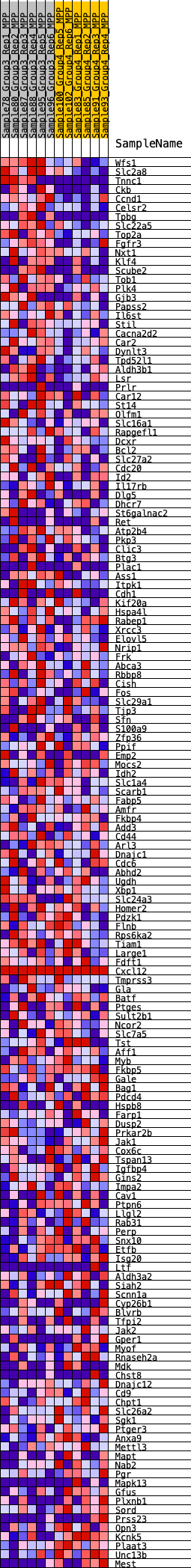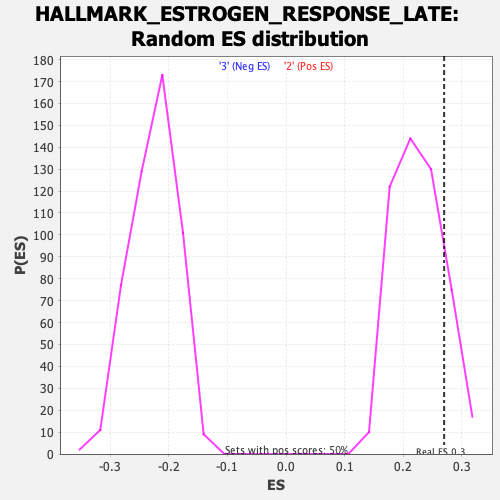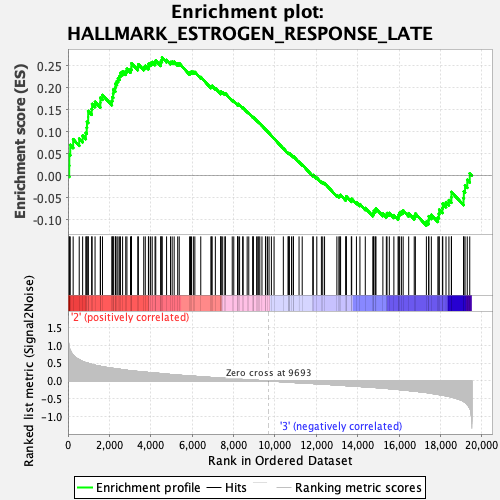
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_ESTROGEN_RESPONSE_LATE |
| Enrichment Score (ES) | 0.2695493 |
| Normalized Enrichment Score (NES) | 1.1974238 |
| Nominal p-value | 0.16666667 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.948 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Wfs1 | 58 | 0.927 | 0.0229 | Yes |
| 2 | Slc2a8 | 63 | 0.922 | 0.0484 | Yes |
| 3 | Tnnc1 | 108 | 0.858 | 0.0701 | Yes |
| 4 | Ckb | 249 | 0.721 | 0.0830 | Yes |
| 5 | Ccnd1 | 542 | 0.592 | 0.0845 | Yes |
| 6 | Celsr2 | 707 | 0.543 | 0.0912 | Yes |
| 7 | Tpbg | 861 | 0.511 | 0.0976 | Yes |
| 8 | Slc22a5 | 906 | 0.504 | 0.1093 | Yes |
| 9 | Top2a | 917 | 0.502 | 0.1229 | Yes |
| 10 | Fgfr3 | 974 | 0.492 | 0.1337 | Yes |
| 11 | Nxt1 | 975 | 0.492 | 0.1474 | Yes |
| 12 | Klf4 | 1143 | 0.462 | 0.1517 | Yes |
| 13 | Scube2 | 1165 | 0.460 | 0.1635 | Yes |
| 14 | Tob1 | 1307 | 0.437 | 0.1684 | Yes |
| 15 | Plk4 | 1563 | 0.408 | 0.1666 | Yes |
| 16 | Gjb3 | 1566 | 0.408 | 0.1779 | Yes |
| 17 | Papss2 | 1666 | 0.396 | 0.1838 | Yes |
| 18 | Il6st | 2119 | 0.354 | 0.1704 | Yes |
| 19 | Stil | 2142 | 0.352 | 0.1791 | Yes |
| 20 | Cacna2d2 | 2181 | 0.350 | 0.1869 | Yes |
| 21 | Car2 | 2187 | 0.349 | 0.1964 | Yes |
| 22 | Dynlt3 | 2285 | 0.340 | 0.2009 | Yes |
| 23 | Tpd52l1 | 2294 | 0.339 | 0.2099 | Yes |
| 24 | Aldh3b1 | 2364 | 0.332 | 0.2156 | Yes |
| 25 | Lsr | 2417 | 0.328 | 0.2221 | Yes |
| 26 | Prlr | 2496 | 0.325 | 0.2271 | Yes |
| 27 | Car12 | 2534 | 0.322 | 0.2342 | Yes |
| 28 | St14 | 2638 | 0.314 | 0.2377 | Yes |
| 29 | Olfm1 | 2790 | 0.301 | 0.2383 | Yes |
| 30 | Slc16a1 | 2847 | 0.296 | 0.2436 | Yes |
| 31 | Rapgefl1 | 3020 | 0.285 | 0.2427 | Yes |
| 32 | Dcxr | 3066 | 0.282 | 0.2483 | Yes |
| 33 | Bcl2 | 3067 | 0.282 | 0.2561 | Yes |
| 34 | Slc27a2 | 3373 | 0.264 | 0.2477 | Yes |
| 35 | Cdc20 | 3399 | 0.262 | 0.2538 | Yes |
| 36 | Id2 | 3660 | 0.249 | 0.2473 | Yes |
| 37 | Il17rb | 3734 | 0.245 | 0.2504 | Yes |
| 38 | Dlg5 | 3886 | 0.235 | 0.2491 | Yes |
| 39 | Dhcr7 | 3902 | 0.234 | 0.2549 | Yes |
| 40 | St6galnac2 | 3991 | 0.229 | 0.2568 | Yes |
| 41 | Ret | 4069 | 0.225 | 0.2590 | Yes |
| 42 | Atp2b4 | 4206 | 0.218 | 0.2581 | Yes |
| 43 | Pkp3 | 4240 | 0.217 | 0.2625 | Yes |
| 44 | Clic3 | 4480 | 0.205 | 0.2559 | Yes |
| 45 | Btg3 | 4490 | 0.205 | 0.2611 | Yes |
| 46 | Plac1 | 4536 | 0.202 | 0.2644 | Yes |
| 47 | Ass1 | 4547 | 0.201 | 0.2695 | Yes |
| 48 | Itpk1 | 4763 | 0.189 | 0.2637 | No |
| 49 | Cdh1 | 4953 | 0.180 | 0.2590 | No |
| 50 | Kif20a | 5033 | 0.176 | 0.2598 | No |
| 51 | Hspa4l | 5130 | 0.171 | 0.2597 | No |
| 52 | Rabep1 | 5301 | 0.163 | 0.2554 | No |
| 53 | Xrcc3 | 5374 | 0.159 | 0.2561 | No |
| 54 | Elovl5 | 5876 | 0.141 | 0.2342 | No |
| 55 | Nrip1 | 5920 | 0.138 | 0.2358 | No |
| 56 | Frk | 5966 | 0.137 | 0.2373 | No |
| 57 | Abca3 | 6057 | 0.133 | 0.2364 | No |
| 58 | Rbbp8 | 6129 | 0.129 | 0.2363 | No |
| 59 | Cish | 6415 | 0.117 | 0.2248 | No |
| 60 | Fos | 6904 | 0.097 | 0.2023 | No |
| 61 | Slc29a1 | 6942 | 0.096 | 0.2031 | No |
| 62 | Tjp3 | 6963 | 0.095 | 0.2047 | No |
| 63 | Sfn | 7120 | 0.090 | 0.1992 | No |
| 64 | S100a9 | 7378 | 0.080 | 0.1881 | No |
| 65 | Zfp36 | 7393 | 0.079 | 0.1896 | No |
| 66 | Ppif | 7395 | 0.079 | 0.1918 | No |
| 67 | Emp2 | 7475 | 0.077 | 0.1898 | No |
| 68 | Mocs2 | 7581 | 0.073 | 0.1865 | No |
| 69 | Idh2 | 7605 | 0.073 | 0.1873 | No |
| 70 | Slc1a4 | 7936 | 0.061 | 0.1720 | No |
| 71 | Scarb1 | 8010 | 0.058 | 0.1698 | No |
| 72 | Fabp5 | 8192 | 0.052 | 0.1619 | No |
| 73 | Amfr | 8199 | 0.051 | 0.1630 | No |
| 74 | Fkbp4 | 8219 | 0.051 | 0.1635 | No |
| 75 | Add3 | 8291 | 0.048 | 0.1611 | No |
| 76 | Cd44 | 8439 | 0.042 | 0.1547 | No |
| 77 | Arl3 | 8463 | 0.041 | 0.1546 | No |
| 78 | Dnajc1 | 8657 | 0.034 | 0.1456 | No |
| 79 | Cdc6 | 8735 | 0.031 | 0.1425 | No |
| 80 | Abhd2 | 8926 | 0.025 | 0.1334 | No |
| 81 | Ugdh | 8942 | 0.024 | 0.1333 | No |
| 82 | Xbp1 | 8954 | 0.023 | 0.1334 | No |
| 83 | Slc24a3 | 9114 | 0.018 | 0.1257 | No |
| 84 | Homer2 | 9170 | 0.016 | 0.1233 | No |
| 85 | Pdzk1 | 9216 | 0.014 | 0.1214 | No |
| 86 | Flnb | 9268 | 0.013 | 0.1191 | No |
| 87 | Rps6ka2 | 9372 | 0.009 | 0.1140 | No |
| 88 | Tiam1 | 9541 | 0.005 | 0.1055 | No |
| 89 | Large1 | 9557 | 0.004 | 0.1048 | No |
| 90 | Fdft1 | 9643 | 0.001 | 0.1005 | No |
| 91 | Cxcl12 | 9715 | 0.000 | 0.0968 | No |
| 92 | Tmprss3 | 9828 | -0.002 | 0.0911 | No |
| 93 | Gla | 9957 | -0.007 | 0.0846 | No |
| 94 | Batf | 10405 | -0.022 | 0.0622 | No |
| 95 | Ptges | 10638 | -0.030 | 0.0510 | No |
| 96 | Sult2b1 | 10644 | -0.030 | 0.0516 | No |
| 97 | Ncor2 | 10689 | -0.032 | 0.0502 | No |
| 98 | Slc7a5 | 10691 | -0.032 | 0.0510 | No |
| 99 | Tst | 10800 | -0.036 | 0.0465 | No |
| 100 | Aff1 | 10882 | -0.039 | 0.0434 | No |
| 101 | Myb | 10893 | -0.040 | 0.0440 | No |
| 102 | Fkbp5 | 11164 | -0.050 | 0.0314 | No |
| 103 | Gale | 11317 | -0.055 | 0.0251 | No |
| 104 | Bag1 | 11828 | -0.070 | 0.0007 | No |
| 105 | Pdcd4 | 11852 | -0.071 | 0.0015 | No |
| 106 | Hspb8 | 12022 | -0.077 | -0.0051 | No |
| 107 | Farp1 | 12244 | -0.084 | -0.0141 | No |
| 108 | Dusp2 | 12309 | -0.086 | -0.0150 | No |
| 109 | Prkar2b | 12397 | -0.090 | -0.0170 | No |
| 110 | Jak1 | 12992 | -0.111 | -0.0446 | No |
| 111 | Cox6c | 13089 | -0.115 | -0.0464 | No |
| 112 | Tspan13 | 13133 | -0.117 | -0.0453 | No |
| 113 | Igfbp4 | 13171 | -0.118 | -0.0439 | No |
| 114 | Gins2 | 13419 | -0.127 | -0.0531 | No |
| 115 | Impa2 | 13443 | -0.128 | -0.0507 | No |
| 116 | Cav1 | 13445 | -0.128 | -0.0472 | No |
| 117 | Ptpn6 | 13690 | -0.138 | -0.0559 | No |
| 118 | Llgl2 | 13708 | -0.139 | -0.0529 | No |
| 119 | Rab31 | 13937 | -0.146 | -0.0606 | No |
| 120 | Perp | 14106 | -0.153 | -0.0650 | No |
| 121 | Snx10 | 14365 | -0.163 | -0.0738 | No |
| 122 | Etfb | 14729 | -0.179 | -0.0876 | No |
| 123 | Isg20 | 14763 | -0.180 | -0.0842 | No |
| 124 | Ltf | 14782 | -0.180 | -0.0801 | No |
| 125 | Aldh3a2 | 14852 | -0.181 | -0.0786 | No |
| 126 | Siah2 | 14881 | -0.183 | -0.0750 | No |
| 127 | Scnn1a | 15215 | -0.198 | -0.0867 | No |
| 128 | Cyp26b1 | 15382 | -0.206 | -0.0895 | No |
| 129 | Blvrb | 15414 | -0.207 | -0.0853 | No |
| 130 | Tfpi2 | 15526 | -0.213 | -0.0851 | No |
| 131 | Jak2 | 15746 | -0.224 | -0.0901 | No |
| 132 | Gper1 | 15949 | -0.236 | -0.0940 | No |
| 133 | Myof | 15981 | -0.238 | -0.0889 | No |
| 134 | Rnaseh2a | 16024 | -0.240 | -0.0844 | No |
| 135 | Mdk | 16110 | -0.245 | -0.0820 | No |
| 136 | Chst8 | 16195 | -0.250 | -0.0793 | No |
| 137 | Dnajc12 | 16462 | -0.265 | -0.0857 | No |
| 138 | Cd9 | 16730 | -0.282 | -0.0916 | No |
| 139 | Chpt1 | 16785 | -0.285 | -0.0864 | No |
| 140 | Slc26a2 | 17317 | -0.320 | -0.1049 | No |
| 141 | Sgk1 | 17428 | -0.328 | -0.1014 | No |
| 142 | Ptger3 | 17434 | -0.328 | -0.0925 | No |
| 143 | Anxa9 | 17557 | -0.342 | -0.0893 | No |
| 144 | Mettl3 | 17873 | -0.372 | -0.0952 | No |
| 145 | Mapt | 17920 | -0.378 | -0.0870 | No |
| 146 | Nab2 | 17939 | -0.379 | -0.0773 | No |
| 147 | Pgr | 18091 | -0.393 | -0.0741 | No |
| 148 | Mapk13 | 18109 | -0.396 | -0.0640 | No |
| 149 | Gfus | 18265 | -0.412 | -0.0605 | No |
| 150 | Plxnb1 | 18406 | -0.428 | -0.0558 | No |
| 151 | Sord | 18524 | -0.445 | -0.0494 | No |
| 152 | Prss23 | 18530 | -0.445 | -0.0372 | No |
| 153 | Opn3 | 19111 | -0.569 | -0.0513 | No |
| 154 | Kcnk5 | 19127 | -0.572 | -0.0361 | No |
| 155 | Plaat3 | 19185 | -0.597 | -0.0224 | No |
| 156 | Unc13b | 19292 | -0.656 | -0.0095 | No |
| 157 | Mest | 19412 | -0.747 | 0.0052 | No |