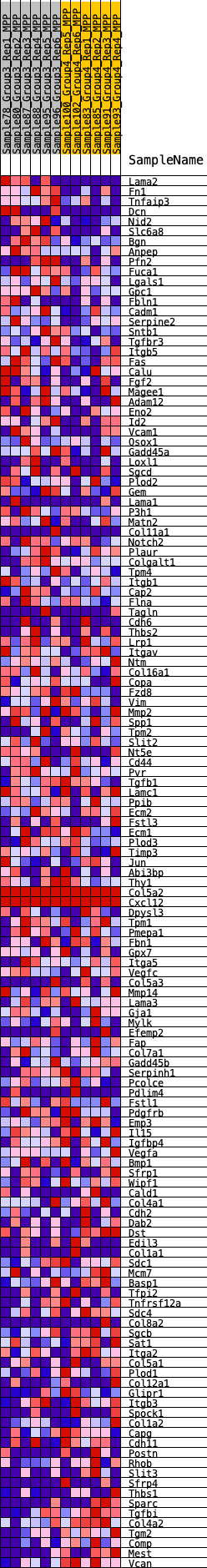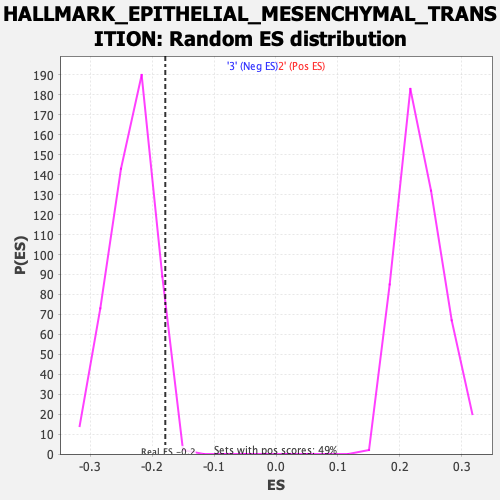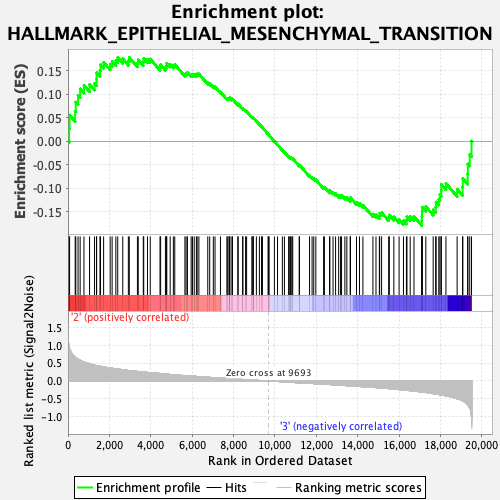
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | -0.17895634 |
| Normalized Enrichment Score (NES) | -0.76838094 |
| Nominal p-value | 0.9726027 |
| FDR q-value | 0.960786 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Lama2 | 53 | 0.938 | 0.0278 | Yes |
| 2 | Fn1 | 77 | 0.900 | 0.0560 | Yes |
| 3 | Tnfaip3 | 347 | 0.666 | 0.0638 | Yes |
| 4 | Dcn | 377 | 0.652 | 0.0836 | Yes |
| 5 | Nid2 | 486 | 0.610 | 0.0979 | Yes |
| 6 | Slc6a8 | 587 | 0.578 | 0.1116 | Yes |
| 7 | Bgn | 771 | 0.531 | 0.1195 | Yes |
| 8 | Anpep | 1042 | 0.477 | 0.1211 | Yes |
| 9 | Pfn2 | 1282 | 0.439 | 0.1230 | Yes |
| 10 | Fuca1 | 1379 | 0.429 | 0.1321 | Yes |
| 11 | Lgals1 | 1382 | 0.429 | 0.1459 | Yes |
| 12 | Gpc1 | 1545 | 0.410 | 0.1510 | Yes |
| 13 | Fbln1 | 1572 | 0.407 | 0.1629 | Yes |
| 14 | Cadm1 | 1721 | 0.390 | 0.1680 | Yes |
| 15 | Serpine2 | 2038 | 0.361 | 0.1634 | Yes |
| 16 | Sntb1 | 2132 | 0.353 | 0.1701 | Yes |
| 17 | Tgfbr3 | 2312 | 0.337 | 0.1718 | Yes |
| 18 | Itgb5 | 2404 | 0.328 | 0.1778 | Yes |
| 19 | Fas | 2646 | 0.313 | 0.1756 | Yes |
| 20 | Calu | 2916 | 0.291 | 0.1712 | Yes |
| 21 | Fgf2 | 2961 | 0.289 | 0.1784 | Yes |
| 22 | Magee1 | 3355 | 0.265 | 0.1667 | No |
| 23 | Adam12 | 3394 | 0.262 | 0.1733 | No |
| 24 | Eno2 | 3638 | 0.250 | 0.1689 | No |
| 25 | Id2 | 3660 | 0.249 | 0.1759 | No |
| 26 | Vcam1 | 3840 | 0.238 | 0.1744 | No |
| 27 | Qsox1 | 3975 | 0.230 | 0.1750 | No |
| 28 | Gadd45a | 4446 | 0.207 | 0.1575 | No |
| 29 | Loxl1 | 4476 | 0.206 | 0.1627 | No |
| 30 | Sgcd | 4708 | 0.193 | 0.1571 | No |
| 31 | Plod2 | 4754 | 0.190 | 0.1610 | No |
| 32 | Gem | 4773 | 0.189 | 0.1662 | No |
| 33 | Lama1 | 4937 | 0.180 | 0.1637 | No |
| 34 | P3h1 | 5090 | 0.173 | 0.1615 | No |
| 35 | Matn2 | 5158 | 0.169 | 0.1635 | No |
| 36 | Col11a1 | 5649 | 0.148 | 0.1431 | No |
| 37 | Notch2 | 5707 | 0.148 | 0.1450 | No |
| 38 | Plaur | 5772 | 0.145 | 0.1464 | No |
| 39 | Colgalt1 | 5951 | 0.137 | 0.1417 | No |
| 40 | Tpm4 | 6014 | 0.134 | 0.1428 | No |
| 41 | Itgb1 | 6099 | 0.131 | 0.1428 | No |
| 42 | Cap2 | 6204 | 0.125 | 0.1415 | No |
| 43 | Flna | 6240 | 0.124 | 0.1437 | No |
| 44 | Tagln | 6324 | 0.120 | 0.1433 | No |
| 45 | Cdh6 | 6754 | 0.103 | 0.1245 | No |
| 46 | Thbs2 | 6847 | 0.099 | 0.1230 | No |
| 47 | Lrp1 | 7021 | 0.093 | 0.1171 | No |
| 48 | Itgav | 7104 | 0.090 | 0.1158 | No |
| 49 | Ntm | 7366 | 0.080 | 0.1050 | No |
| 50 | Col16a1 | 7677 | 0.071 | 0.0913 | No |
| 51 | Copa | 7719 | 0.069 | 0.0914 | No |
| 52 | Fzd8 | 7781 | 0.066 | 0.0904 | No |
| 53 | Vim | 7817 | 0.065 | 0.0907 | No |
| 54 | Mmp2 | 7818 | 0.065 | 0.0929 | No |
| 55 | Spp1 | 7921 | 0.061 | 0.0896 | No |
| 56 | Tpm2 | 7947 | 0.060 | 0.0903 | No |
| 57 | Slit2 | 8207 | 0.051 | 0.0785 | No |
| 58 | Nt5e | 8220 | 0.051 | 0.0796 | No |
| 59 | Cd44 | 8439 | 0.042 | 0.0697 | No |
| 60 | Pvr | 8450 | 0.041 | 0.0705 | No |
| 61 | Tgfb1 | 8581 | 0.037 | 0.0650 | No |
| 62 | Lamc1 | 8630 | 0.035 | 0.0637 | No |
| 63 | Ppib | 8884 | 0.026 | 0.0514 | No |
| 64 | Ecm2 | 8940 | 0.024 | 0.0494 | No |
| 65 | Fstl3 | 8941 | 0.024 | 0.0502 | No |
| 66 | Ecm1 | 8947 | 0.024 | 0.0507 | No |
| 67 | Plod3 | 9102 | 0.019 | 0.0433 | No |
| 68 | Timp3 | 9248 | 0.013 | 0.0363 | No |
| 69 | Jun | 9351 | 0.010 | 0.0313 | No |
| 70 | Abi3bp | 9395 | 0.009 | 0.0294 | No |
| 71 | Thy1 | 9666 | 0.001 | 0.0155 | No |
| 72 | Col5a2 | 9698 | 0.000 | 0.0139 | No |
| 73 | Cxcl12 | 9715 | 0.000 | 0.0131 | No |
| 74 | Dpysl3 | 9973 | -0.007 | 0.0000 | No |
| 75 | Tpm1 | 10124 | -0.012 | -0.0073 | No |
| 76 | Pmepa1 | 10358 | -0.020 | -0.0187 | No |
| 77 | Fbn1 | 10456 | -0.024 | -0.0229 | No |
| 78 | Gpx7 | 10660 | -0.031 | -0.0324 | No |
| 79 | Itga5 | 10692 | -0.032 | -0.0329 | No |
| 80 | Vegfc | 10758 | -0.035 | -0.0351 | No |
| 81 | Col5a3 | 10771 | -0.035 | -0.0346 | No |
| 82 | Mmp14 | 10823 | -0.037 | -0.0360 | No |
| 83 | Lama3 | 10859 | -0.038 | -0.0366 | No |
| 84 | Gja1 | 11171 | -0.050 | -0.0510 | No |
| 85 | Mylk | 11185 | -0.050 | -0.0501 | No |
| 86 | Efemp2 | 11671 | -0.066 | -0.0729 | No |
| 87 | Fap | 11794 | -0.069 | -0.0770 | No |
| 88 | Col7a1 | 11874 | -0.072 | -0.0787 | No |
| 89 | Gadd45b | 11976 | -0.075 | -0.0815 | No |
| 90 | Serpinh1 | 12351 | -0.088 | -0.0979 | No |
| 91 | Pcolce | 12390 | -0.090 | -0.0969 | No |
| 92 | Pdlim4 | 12641 | -0.098 | -0.1067 | No |
| 93 | Fstl1 | 12650 | -0.098 | -0.1039 | No |
| 94 | Pdgfrb | 12807 | -0.104 | -0.1086 | No |
| 95 | Emp3 | 12930 | -0.109 | -0.1113 | No |
| 96 | Il15 | 13079 | -0.115 | -0.1152 | No |
| 97 | Igfbp4 | 13171 | -0.118 | -0.1161 | No |
| 98 | Vegfa | 13218 | -0.120 | -0.1145 | No |
| 99 | Bmp1 | 13378 | -0.126 | -0.1186 | No |
| 100 | Sfrp1 | 13474 | -0.130 | -0.1193 | No |
| 101 | Wipf1 | 13634 | -0.136 | -0.1231 | No |
| 102 | Cald1 | 13650 | -0.137 | -0.1194 | No |
| 103 | Col4a1 | 13940 | -0.146 | -0.1295 | No |
| 104 | Cdh2 | 14084 | -0.152 | -0.1320 | No |
| 105 | Dab2 | 14250 | -0.159 | -0.1353 | No |
| 106 | Dst | 14734 | -0.179 | -0.1544 | No |
| 107 | Edil3 | 14882 | -0.183 | -0.1560 | No |
| 108 | Col1a1 | 15044 | -0.189 | -0.1582 | No |
| 109 | Sdc1 | 15060 | -0.190 | -0.1528 | No |
| 110 | Mcm7 | 15153 | -0.194 | -0.1512 | No |
| 111 | Basp1 | 15495 | -0.212 | -0.1619 | No |
| 112 | Tfpi2 | 15526 | -0.213 | -0.1565 | No |
| 113 | Tnfrsf12a | 15745 | -0.224 | -0.1604 | No |
| 114 | Sdc4 | 15998 | -0.239 | -0.1657 | No |
| 115 | Col8a2 | 16209 | -0.250 | -0.1683 | No |
| 116 | Sgcb | 16352 | -0.258 | -0.1673 | No |
| 117 | Sat1 | 16378 | -0.260 | -0.1601 | No |
| 118 | Itga2 | 16535 | -0.270 | -0.1593 | No |
| 119 | Col5a1 | 16718 | -0.281 | -0.1595 | No |
| 120 | Plod1 | 17095 | -0.306 | -0.1690 | No |
| 121 | Col12a1 | 17099 | -0.306 | -0.1592 | No |
| 122 | Glipr1 | 17107 | -0.307 | -0.1495 | No |
| 123 | Itgb3 | 17113 | -0.307 | -0.1398 | No |
| 124 | Spock1 | 17290 | -0.318 | -0.1385 | No |
| 125 | Col1a2 | 17645 | -0.352 | -0.1453 | No |
| 126 | Capg | 17768 | -0.362 | -0.1398 | No |
| 127 | Cdh11 | 17786 | -0.364 | -0.1288 | No |
| 128 | Postn | 17916 | -0.377 | -0.1232 | No |
| 129 | Rhob | 17970 | -0.383 | -0.1134 | No |
| 130 | Slit3 | 18031 | -0.390 | -0.1038 | No |
| 131 | Sfrp4 | 18037 | -0.391 | -0.0914 | No |
| 132 | Thbs1 | 18262 | -0.412 | -0.0895 | No |
| 133 | Sparc | 18802 | -0.493 | -0.1012 | No |
| 134 | Tgfbi | 19066 | -0.552 | -0.0968 | No |
| 135 | Col4a2 | 19077 | -0.556 | -0.0792 | No |
| 136 | Tgm2 | 19313 | -0.670 | -0.0695 | No |
| 137 | Comp | 19322 | -0.676 | -0.0479 | No |
| 138 | Mest | 19412 | -0.747 | -0.0282 | No |
| 139 | Vcan | 19498 | -1.021 | 0.0007 | No |