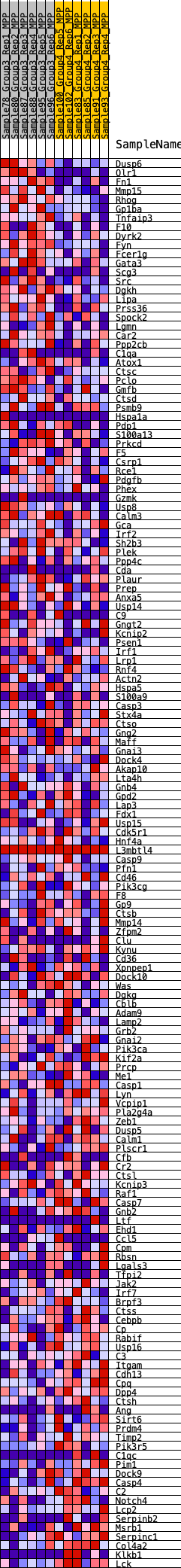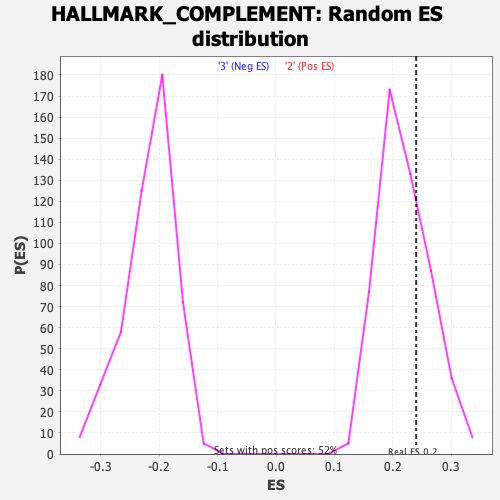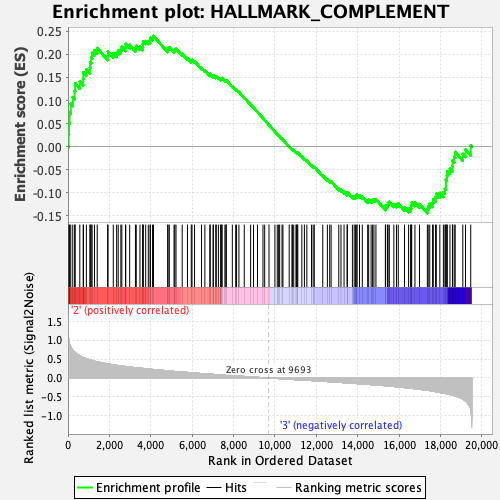
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | 0.24012491 |
| Normalized Enrichment Score (NES) | 1.0948113 |
| Nominal p-value | 0.2947977 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.992 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dusp6 | 30 | 1.050 | 0.0265 | Yes |
| 2 | Olr1 | 47 | 0.956 | 0.0511 | Yes |
| 3 | Fn1 | 77 | 0.900 | 0.0736 | Yes |
| 4 | Mmp15 | 133 | 0.825 | 0.0928 | Yes |
| 5 | Rhog | 230 | 0.730 | 0.1073 | Yes |
| 6 | Gp1ba | 321 | 0.681 | 0.1208 | Yes |
| 7 | Tnfaip3 | 347 | 0.666 | 0.1373 | Yes |
| 8 | F10 | 570 | 0.585 | 0.1414 | Yes |
| 9 | Dyrk2 | 734 | 0.540 | 0.1474 | Yes |
| 10 | Fyn | 748 | 0.537 | 0.1611 | Yes |
| 11 | Fcer1g | 888 | 0.507 | 0.1674 | Yes |
| 12 | Gata3 | 1061 | 0.475 | 0.1712 | Yes |
| 13 | Scg3 | 1074 | 0.472 | 0.1832 | Yes |
| 14 | Src | 1122 | 0.465 | 0.1932 | Yes |
| 15 | Dgkh | 1164 | 0.460 | 0.2033 | Yes |
| 16 | Lipa | 1279 | 0.440 | 0.2092 | Yes |
| 17 | Prss36 | 1412 | 0.425 | 0.2137 | Yes |
| 18 | Spock2 | 1917 | 0.371 | 0.1975 | Yes |
| 19 | Lgmn | 1933 | 0.369 | 0.2066 | Yes |
| 20 | Car2 | 2187 | 0.349 | 0.2029 | Yes |
| 21 | Ppp2cb | 2351 | 0.333 | 0.2033 | Yes |
| 22 | C1qa | 2426 | 0.327 | 0.2082 | Yes |
| 23 | Atox1 | 2554 | 0.320 | 0.2102 | Yes |
| 24 | Ctsc | 2601 | 0.317 | 0.2163 | Yes |
| 25 | Pclo | 2778 | 0.302 | 0.2152 | Yes |
| 26 | Gmfb | 2791 | 0.301 | 0.2227 | Yes |
| 27 | Ctsd | 2980 | 0.288 | 0.2206 | Yes |
| 28 | Psmb9 | 3259 | 0.270 | 0.2135 | Yes |
| 29 | Hspa1a | 3297 | 0.268 | 0.2187 | Yes |
| 30 | Pdp1 | 3477 | 0.259 | 0.2164 | Yes |
| 31 | S100a13 | 3610 | 0.252 | 0.2163 | Yes |
| 32 | Prkcd | 3619 | 0.251 | 0.2225 | Yes |
| 33 | F5 | 3635 | 0.250 | 0.2284 | Yes |
| 34 | Csrp1 | 3753 | 0.243 | 0.2289 | Yes |
| 35 | Rce1 | 3880 | 0.236 | 0.2287 | Yes |
| 36 | Pdgfb | 3968 | 0.231 | 0.2303 | Yes |
| 37 | Phex | 3978 | 0.230 | 0.2360 | Yes |
| 38 | Gzmk | 4086 | 0.224 | 0.2364 | Yes |
| 39 | Usp8 | 4131 | 0.223 | 0.2401 | Yes |
| 40 | Calm3 | 4817 | 0.187 | 0.2097 | No |
| 41 | Gca | 4833 | 0.186 | 0.2139 | No |
| 42 | Irf2 | 4895 | 0.183 | 0.2156 | No |
| 43 | Sh2b3 | 5124 | 0.171 | 0.2084 | No |
| 44 | Plek | 5161 | 0.169 | 0.2111 | No |
| 45 | Ppp4c | 5219 | 0.166 | 0.2126 | No |
| 46 | Cda | 5517 | 0.153 | 0.2013 | No |
| 47 | Plaur | 5772 | 0.145 | 0.1921 | No |
| 48 | Prep | 5956 | 0.137 | 0.1863 | No |
| 49 | Anxa5 | 5992 | 0.135 | 0.1881 | No |
| 50 | Usp14 | 6115 | 0.130 | 0.1852 | No |
| 51 | C9 | 6452 | 0.115 | 0.1709 | No |
| 52 | Gngt2 | 6613 | 0.109 | 0.1656 | No |
| 53 | Kcnip2 | 6855 | 0.099 | 0.1557 | No |
| 54 | Psen1 | 6871 | 0.098 | 0.1576 | No |
| 55 | Irf1 | 7013 | 0.093 | 0.1528 | No |
| 56 | Lrp1 | 7021 | 0.093 | 0.1549 | No |
| 57 | Rnf4 | 7141 | 0.089 | 0.1512 | No |
| 58 | Actn2 | 7163 | 0.088 | 0.1524 | No |
| 59 | Hspa5 | 7267 | 0.084 | 0.1493 | No |
| 60 | S100a9 | 7378 | 0.080 | 0.1458 | No |
| 61 | Casp3 | 7392 | 0.079 | 0.1472 | No |
| 62 | Stx4a | 7432 | 0.078 | 0.1473 | No |
| 63 | Ctso | 7444 | 0.078 | 0.1488 | No |
| 64 | Gng2 | 7582 | 0.073 | 0.1437 | No |
| 65 | Maff | 7629 | 0.072 | 0.1432 | No |
| 66 | Gnai3 | 7649 | 0.071 | 0.1442 | No |
| 67 | Dock4 | 7940 | 0.061 | 0.1308 | No |
| 68 | Akap10 | 8104 | 0.055 | 0.1238 | No |
| 69 | Lta4h | 8144 | 0.053 | 0.1232 | No |
| 70 | Gnb4 | 8258 | 0.049 | 0.1187 | No |
| 71 | Gpd2 | 8517 | 0.039 | 0.1064 | No |
| 72 | Lap3 | 8824 | 0.028 | 0.0914 | No |
| 73 | Fdx1 | 8965 | 0.023 | 0.0847 | No |
| 74 | Usp15 | 9156 | 0.017 | 0.0754 | No |
| 75 | Cdk5r1 | 9421 | 0.008 | 0.0619 | No |
| 76 | Hnf4a | 9504 | 0.006 | 0.0579 | No |
| 77 | L3mbtl4 | 9712 | 0.000 | 0.0472 | No |
| 78 | Casp9 | 9998 | -0.008 | 0.0327 | No |
| 79 | Pfn1 | 10112 | -0.012 | 0.0271 | No |
| 80 | Cd46 | 10178 | -0.014 | 0.0242 | No |
| 81 | Pik3cg | 10227 | -0.016 | 0.0221 | No |
| 82 | F8 | 10341 | -0.019 | 0.0168 | No |
| 83 | Gp9 | 10386 | -0.021 | 0.0151 | No |
| 84 | Ctsb | 10688 | -0.032 | 0.0004 | No |
| 85 | Mmp14 | 10823 | -0.037 | -0.0056 | No |
| 86 | Zfpm2 | 10842 | -0.038 | -0.0055 | No |
| 87 | Clu | 10907 | -0.041 | -0.0077 | No |
| 88 | Kynu | 11015 | -0.044 | -0.0120 | No |
| 89 | Cd36 | 11045 | -0.045 | -0.0123 | No |
| 90 | Xpnpep1 | 11073 | -0.047 | -0.0125 | No |
| 91 | Dock10 | 11104 | -0.047 | -0.0128 | No |
| 92 | Was | 11300 | -0.054 | -0.0214 | No |
| 93 | Dgkg | 11423 | -0.058 | -0.0261 | No |
| 94 | Cblb | 11542 | -0.063 | -0.0306 | No |
| 95 | Adam9 | 11766 | -0.068 | -0.0403 | No |
| 96 | Lamp2 | 11844 | -0.071 | -0.0424 | No |
| 97 | Grb2 | 11900 | -0.073 | -0.0433 | No |
| 98 | Gnai2 | 12310 | -0.086 | -0.0621 | No |
| 99 | Pik3ca | 12533 | -0.094 | -0.0710 | No |
| 100 | Kif2a | 12646 | -0.098 | -0.0742 | No |
| 101 | Prcp | 12714 | -0.101 | -0.0750 | No |
| 102 | Me1 | 13084 | -0.115 | -0.0910 | No |
| 103 | Casp1 | 13191 | -0.119 | -0.0933 | No |
| 104 | Lyn | 13339 | -0.124 | -0.0976 | No |
| 105 | Vcpip1 | 13485 | -0.131 | -0.1016 | No |
| 106 | Pla2g4a | 13501 | -0.131 | -0.0989 | No |
| 107 | Zeb1 | 13742 | -0.140 | -0.1075 | No |
| 108 | Dusp5 | 13829 | -0.142 | -0.1082 | No |
| 109 | Calm1 | 13896 | -0.145 | -0.1077 | No |
| 110 | Plscr1 | 13959 | -0.147 | -0.1070 | No |
| 111 | Cfb | 13961 | -0.147 | -0.1031 | No |
| 112 | Cr2 | 14086 | -0.152 | -0.1055 | No |
| 113 | Ctsl | 14221 | -0.157 | -0.1082 | No |
| 114 | Kcnip3 | 14480 | -0.168 | -0.1170 | No |
| 115 | Raf1 | 14529 | -0.170 | -0.1150 | No |
| 116 | Casp7 | 14647 | -0.175 | -0.1164 | No |
| 117 | Gnb2 | 14719 | -0.178 | -0.1153 | No |
| 118 | Ltf | 14782 | -0.180 | -0.1137 | No |
| 119 | Ehd1 | 14879 | -0.183 | -0.1137 | No |
| 120 | Ccl5 | 15335 | -0.204 | -0.1318 | No |
| 121 | Cpm | 15348 | -0.204 | -0.1270 | No |
| 122 | Rbsn | 15440 | -0.209 | -0.1261 | No |
| 123 | Lgals3 | 15464 | -0.210 | -0.1217 | No |
| 124 | Tfpi2 | 15526 | -0.213 | -0.1191 | No |
| 125 | Jak2 | 15746 | -0.224 | -0.1245 | No |
| 126 | Irf7 | 15874 | -0.231 | -0.1249 | No |
| 127 | Brpf3 | 15958 | -0.236 | -0.1229 | No |
| 128 | Ctss | 16260 | -0.253 | -0.1317 | No |
| 129 | Cebpb | 16458 | -0.265 | -0.1348 | No |
| 130 | Cp | 16555 | -0.271 | -0.1325 | No |
| 131 | Rabif | 16591 | -0.273 | -0.1271 | No |
| 132 | Usp16 | 16607 | -0.275 | -0.1205 | No |
| 133 | C3 | 16765 | -0.284 | -0.1210 | No |
| 134 | Itgam | 16984 | -0.299 | -0.1243 | No |
| 135 | Cdh13 | 17364 | -0.325 | -0.1353 | No |
| 136 | Cpq | 17404 | -0.327 | -0.1286 | No |
| 137 | Dpp4 | 17479 | -0.333 | -0.1235 | No |
| 138 | Ctsh | 17617 | -0.349 | -0.1213 | No |
| 139 | Ang | 17649 | -0.352 | -0.1135 | No |
| 140 | Sirt6 | 17761 | -0.361 | -0.1096 | No |
| 141 | Prdm4 | 17796 | -0.365 | -0.1016 | No |
| 142 | Timp2 | 17967 | -0.383 | -0.1002 | No |
| 143 | Pik3r5 | 18139 | -0.398 | -0.0984 | No |
| 144 | C1qc | 18213 | -0.406 | -0.0913 | No |
| 145 | Pim1 | 18259 | -0.412 | -0.0827 | No |
| 146 | Dock9 | 18260 | -0.412 | -0.0717 | No |
| 147 | Casp4 | 18308 | -0.417 | -0.0630 | No |
| 148 | C2 | 18326 | -0.419 | -0.0527 | No |
| 149 | Notch4 | 18456 | -0.436 | -0.0477 | No |
| 150 | Lcp2 | 18576 | -0.451 | -0.0418 | No |
| 151 | Serpinb2 | 18583 | -0.452 | -0.0301 | No |
| 152 | Msrb1 | 18661 | -0.466 | -0.0216 | No |
| 153 | Serpinc1 | 18709 | -0.473 | -0.0115 | No |
| 154 | Col4a2 | 19077 | -0.556 | -0.0156 | No |
| 155 | Klkb1 | 19207 | -0.605 | -0.0061 | No |
| 156 | Lck | 19460 | -0.817 | 0.0027 | No |