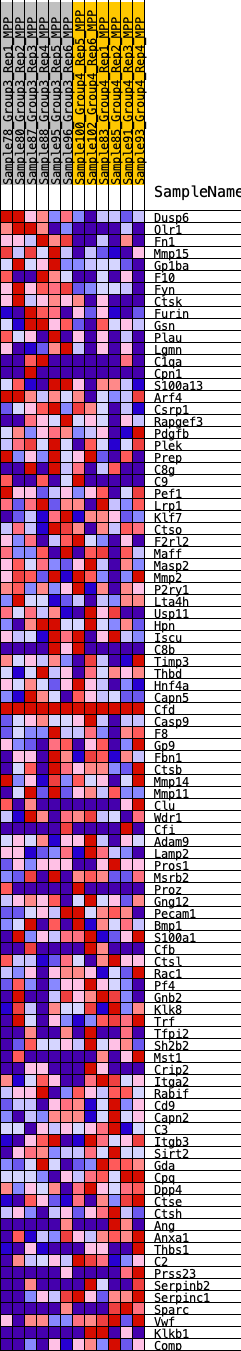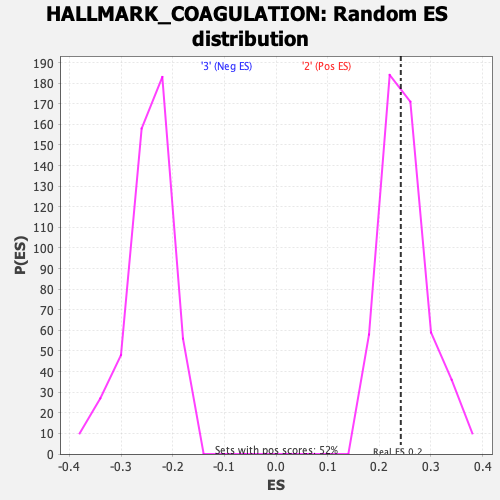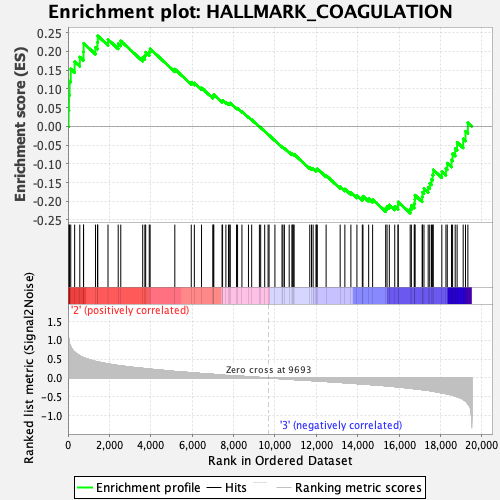
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.24155413 |
| Normalized Enrichment Score (NES) | 0.9680432 |
| Nominal p-value | 0.52509654 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dusp6 | 30 | 1.050 | 0.0435 | Yes |
| 2 | Olr1 | 47 | 0.956 | 0.0837 | Yes |
| 3 | Fn1 | 77 | 0.900 | 0.1208 | Yes |
| 4 | Mmp15 | 133 | 0.825 | 0.1533 | Yes |
| 5 | Gp1ba | 321 | 0.681 | 0.1729 | Yes |
| 6 | F10 | 570 | 0.585 | 0.1852 | Yes |
| 7 | Fyn | 748 | 0.537 | 0.1991 | Yes |
| 8 | Ctsk | 754 | 0.536 | 0.2218 | Yes |
| 9 | Furin | 1328 | 0.434 | 0.2109 | Yes |
| 10 | Gsn | 1424 | 0.424 | 0.2242 | Yes |
| 11 | Plau | 1439 | 0.422 | 0.2416 | Yes |
| 12 | Lgmn | 1933 | 0.369 | 0.2320 | No |
| 13 | C1qa | 2426 | 0.327 | 0.2207 | No |
| 14 | Cpn1 | 2550 | 0.321 | 0.2281 | No |
| 15 | S100a13 | 3610 | 0.252 | 0.1844 | No |
| 16 | Arf4 | 3715 | 0.246 | 0.1896 | No |
| 17 | Csrp1 | 3753 | 0.243 | 0.1981 | No |
| 18 | Rapgef3 | 3926 | 0.233 | 0.1992 | No |
| 19 | Pdgfb | 3968 | 0.231 | 0.2070 | No |
| 20 | Plek | 5161 | 0.169 | 0.1529 | No |
| 21 | Prep | 5956 | 0.137 | 0.1178 | No |
| 22 | C8g | 6105 | 0.130 | 0.1158 | No |
| 23 | C9 | 6452 | 0.115 | 0.1029 | No |
| 24 | Pef1 | 6998 | 0.094 | 0.0789 | No |
| 25 | Lrp1 | 7021 | 0.093 | 0.0817 | No |
| 26 | Klf7 | 7046 | 0.092 | 0.0845 | No |
| 27 | Ctso | 7444 | 0.078 | 0.0674 | No |
| 28 | F2rl2 | 7469 | 0.077 | 0.0694 | No |
| 29 | Maff | 7629 | 0.072 | 0.0643 | No |
| 30 | Masp2 | 7754 | 0.067 | 0.0608 | No |
| 31 | Mmp2 | 7818 | 0.065 | 0.0604 | No |
| 32 | P2ry1 | 7835 | 0.064 | 0.0623 | No |
| 33 | Lta4h | 8144 | 0.053 | 0.0487 | No |
| 34 | Usp11 | 8193 | 0.052 | 0.0484 | No |
| 35 | Hpn | 8404 | 0.043 | 0.0395 | No |
| 36 | Iscu | 8720 | 0.032 | 0.0246 | No |
| 37 | C8b | 8875 | 0.026 | 0.0178 | No |
| 38 | Timp3 | 9248 | 0.013 | -0.0008 | No |
| 39 | Thbd | 9310 | 0.011 | -0.0034 | No |
| 40 | Hnf4a | 9504 | 0.006 | -0.0131 | No |
| 41 | Capn5 | 9668 | 0.001 | -0.0215 | No |
| 42 | Cfd | 9716 | 0.000 | -0.0239 | No |
| 43 | Casp9 | 9998 | -0.008 | -0.0380 | No |
| 44 | F8 | 10341 | -0.019 | -0.0548 | No |
| 45 | Gp9 | 10386 | -0.021 | -0.0562 | No |
| 46 | Fbn1 | 10456 | -0.024 | -0.0587 | No |
| 47 | Ctsb | 10688 | -0.032 | -0.0692 | No |
| 48 | Mmp14 | 10823 | -0.037 | -0.0745 | No |
| 49 | Mmp11 | 10839 | -0.038 | -0.0737 | No |
| 50 | Clu | 10907 | -0.041 | -0.0754 | No |
| 51 | Wdr1 | 10921 | -0.041 | -0.0743 | No |
| 52 | Cfi | 11682 | -0.066 | -0.1106 | No |
| 53 | Adam9 | 11766 | -0.068 | -0.1120 | No |
| 54 | Lamp2 | 11844 | -0.071 | -0.1129 | No |
| 55 | Pros1 | 11982 | -0.075 | -0.1167 | No |
| 56 | Msrb2 | 12014 | -0.077 | -0.1150 | No |
| 57 | Proz | 12045 | -0.078 | -0.1132 | No |
| 58 | Gng12 | 12472 | -0.092 | -0.1312 | No |
| 59 | Pecam1 | 13151 | -0.117 | -0.1611 | No |
| 60 | Bmp1 | 13378 | -0.126 | -0.1673 | No |
| 61 | S100a1 | 13669 | -0.138 | -0.1763 | No |
| 62 | Cfb | 13961 | -0.147 | -0.1850 | No |
| 63 | Ctsl | 14221 | -0.157 | -0.1916 | No |
| 64 | Rac1 | 14252 | -0.159 | -0.1863 | No |
| 65 | Pf4 | 14528 | -0.170 | -0.1932 | No |
| 66 | Gnb2 | 14719 | -0.178 | -0.1954 | No |
| 67 | Klk8 | 15350 | -0.204 | -0.2190 | No |
| 68 | Trf | 15416 | -0.208 | -0.2135 | No |
| 69 | Tfpi2 | 15526 | -0.213 | -0.2099 | No |
| 70 | Sh2b2 | 15788 | -0.227 | -0.2137 | No |
| 71 | Mst1 | 15948 | -0.236 | -0.2118 | No |
| 72 | Crip2 | 15952 | -0.236 | -0.2018 | No |
| 73 | Itga2 | 16535 | -0.270 | -0.2202 | No |
| 74 | Rabif | 16591 | -0.273 | -0.2113 | No |
| 75 | Cd9 | 16730 | -0.282 | -0.2063 | No |
| 76 | Capn2 | 16759 | -0.284 | -0.1956 | No |
| 77 | C3 | 16765 | -0.284 | -0.1837 | No |
| 78 | Itgb3 | 17113 | -0.307 | -0.1884 | No |
| 79 | Sirt2 | 17129 | -0.308 | -0.1759 | No |
| 80 | Gda | 17200 | -0.312 | -0.1661 | No |
| 81 | Cpq | 17404 | -0.327 | -0.1626 | No |
| 82 | Dpp4 | 17479 | -0.333 | -0.1521 | No |
| 83 | Ctse | 17563 | -0.343 | -0.1417 | No |
| 84 | Ctsh | 17617 | -0.349 | -0.1294 | No |
| 85 | Ang | 17649 | -0.352 | -0.1160 | No |
| 86 | Anxa1 | 18063 | -0.391 | -0.1204 | No |
| 87 | Thbs1 | 18262 | -0.412 | -0.1130 | No |
| 88 | C2 | 18326 | -0.419 | -0.0983 | No |
| 89 | Prss23 | 18530 | -0.445 | -0.0896 | No |
| 90 | Serpinb2 | 18583 | -0.452 | -0.0729 | No |
| 91 | Serpinc1 | 18709 | -0.473 | -0.0591 | No |
| 92 | Sparc | 18802 | -0.493 | -0.0427 | No |
| 93 | Vwf | 19095 | -0.563 | -0.0336 | No |
| 94 | Klkb1 | 19207 | -0.605 | -0.0133 | No |
| 95 | Comp | 19322 | -0.676 | 0.0098 | No |