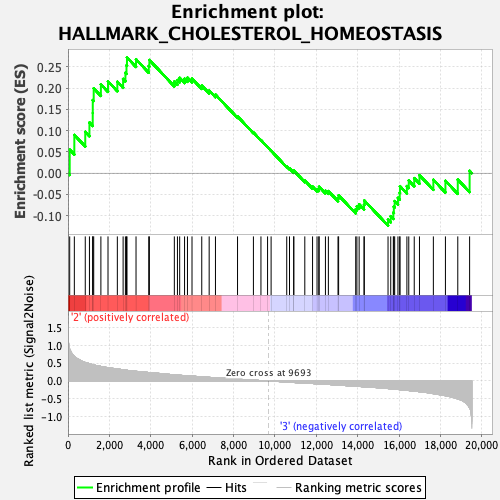
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.27243868 |
| Normalized Enrichment Score (NES) | 0.90497005 |
| Nominal p-value | 0.5666004 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gstm7 | 82 | 0.887 | 0.0553 | Yes |
| 2 | Fbxo6 | 306 | 0.687 | 0.0900 | Yes |
| 3 | Mvk | 834 | 0.514 | 0.0974 | Yes |
| 4 | Ctnnb1 | 1036 | 0.479 | 0.1192 | Yes |
| 5 | Sc5d | 1195 | 0.454 | 0.1416 | Yes |
| 6 | Hmgcs1 | 1197 | 0.454 | 0.1720 | Yes |
| 7 | Hsd17b7 | 1242 | 0.446 | 0.1997 | Yes |
| 8 | Pmvk | 1589 | 0.405 | 0.2090 | Yes |
| 9 | Lgmn | 1933 | 0.369 | 0.2162 | Yes |
| 10 | Sqle | 2384 | 0.329 | 0.2152 | Yes |
| 11 | Lss | 2663 | 0.311 | 0.2218 | Yes |
| 12 | Cyp51 | 2774 | 0.302 | 0.2364 | Yes |
| 13 | Srebf2 | 2827 | 0.298 | 0.2537 | Yes |
| 14 | Errfi1 | 2850 | 0.296 | 0.2724 | Yes |
| 15 | Atf3 | 3287 | 0.268 | 0.2680 | No |
| 16 | Dhcr7 | 3902 | 0.234 | 0.2522 | No |
| 17 | Hmgcr | 3932 | 0.232 | 0.2663 | No |
| 18 | Fasn | 5138 | 0.171 | 0.2158 | No |
| 19 | Niban1 | 5288 | 0.163 | 0.2191 | No |
| 20 | Chka | 5396 | 0.158 | 0.2242 | No |
| 21 | Ldlr | 5632 | 0.149 | 0.2221 | No |
| 22 | Plaur | 5772 | 0.145 | 0.2247 | No |
| 23 | Anxa5 | 5992 | 0.135 | 0.2226 | No |
| 24 | Trib3 | 6463 | 0.115 | 0.2061 | No |
| 25 | Abca2 | 6821 | 0.100 | 0.1945 | No |
| 26 | Mvd | 7125 | 0.089 | 0.1849 | No |
| 27 | Fabp5 | 8192 | 0.052 | 0.1335 | No |
| 28 | Stard4 | 8959 | 0.023 | 0.0957 | No |
| 29 | Pnrc1 | 9323 | 0.011 | 0.0778 | No |
| 30 | Fdft1 | 9643 | 0.001 | 0.0615 | No |
| 31 | Cxcl16 | 9814 | -0.002 | 0.0528 | No |
| 32 | Trp53inp1 | 10569 | -0.028 | 0.0159 | No |
| 33 | Antxr2 | 10703 | -0.032 | 0.0113 | No |
| 34 | Clu | 10907 | -0.041 | 0.0036 | No |
| 35 | Jag1 | 10909 | -0.041 | 0.0063 | No |
| 36 | S100a11 | 11442 | -0.059 | -0.0171 | No |
| 37 | Pcyt2 | 11817 | -0.070 | -0.0317 | No |
| 38 | Fdps | 12033 | -0.077 | -0.0375 | No |
| 39 | Acat2 | 12121 | -0.080 | -0.0366 | No |
| 40 | Acss2 | 12133 | -0.081 | -0.0317 | No |
| 41 | Lpl | 12439 | -0.091 | -0.0413 | No |
| 42 | Atxn2 | 12581 | -0.096 | -0.0421 | No |
| 43 | Gnai1 | 13047 | -0.113 | -0.0584 | No |
| 44 | Actg1 | 13076 | -0.115 | -0.0521 | No |
| 45 | Ech1 | 13902 | -0.145 | -0.0848 | No |
| 46 | Plscr1 | 13959 | -0.147 | -0.0778 | No |
| 47 | Ebp | 14068 | -0.151 | -0.0732 | No |
| 48 | Ethe1 | 14304 | -0.161 | -0.0745 | No |
| 49 | Tmem97 | 14316 | -0.161 | -0.0643 | No |
| 50 | Lgals3 | 15464 | -0.210 | -0.1091 | No |
| 51 | Pdk3 | 15594 | -0.217 | -0.1012 | No |
| 52 | Aldoc | 15720 | -0.223 | -0.0927 | No |
| 53 | Tnfrsf12a | 15745 | -0.224 | -0.0789 | No |
| 54 | Gusb | 15785 | -0.226 | -0.0657 | No |
| 55 | Nfil3 | 15943 | -0.235 | -0.0580 | No |
| 56 | Nsdhl | 16029 | -0.240 | -0.0462 | No |
| 57 | Atf5 | 16041 | -0.241 | -0.0306 | No |
| 58 | Idi1 | 16375 | -0.260 | -0.0303 | No |
| 59 | Alcam | 16466 | -0.265 | -0.0171 | No |
| 60 | Cd9 | 16730 | -0.282 | -0.0117 | No |
| 61 | Cpeb2 | 16983 | -0.299 | -0.0046 | No |
| 62 | Tm7sf2 | 17654 | -0.352 | -0.0154 | No |
| 63 | Stx5a | 18237 | -0.410 | -0.0179 | No |
| 64 | Pparg | 18834 | -0.499 | -0.0150 | No |
| 65 | Fads2 | 19406 | -0.742 | 0.0055 | No |

