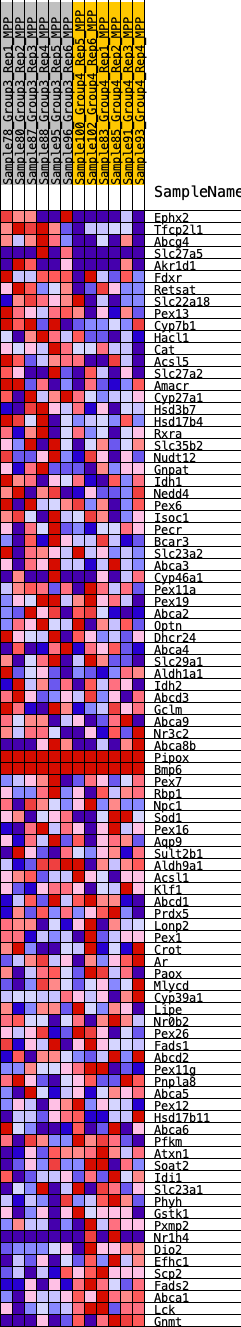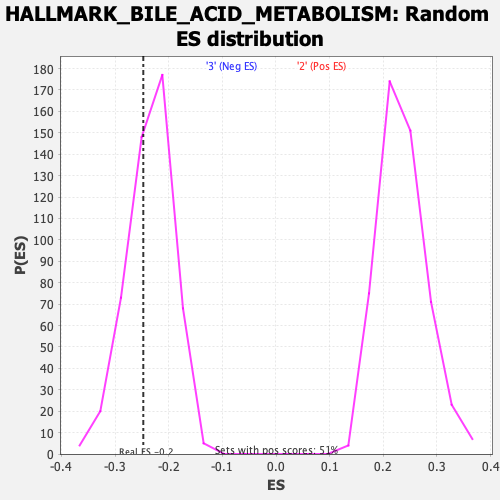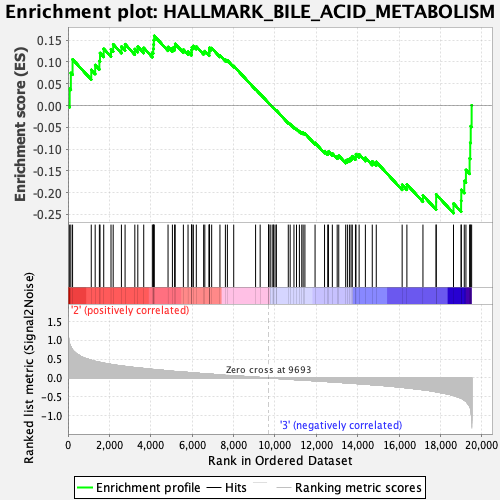
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group3_versus_Group4.MPP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.24731155 |
| Normalized Enrichment Score (NES) | -1.0557745 |
| Nominal p-value | 0.34343433 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ephx2 | 81 | 0.889 | 0.0385 | No |
| 2 | Tfcp2l1 | 138 | 0.820 | 0.0749 | No |
| 3 | Abcg4 | 221 | 0.736 | 0.1060 | No |
| 4 | Slc27a5 | 1124 | 0.465 | 0.0819 | No |
| 5 | Akr1d1 | 1313 | 0.435 | 0.0931 | No |
| 6 | Fdxr | 1516 | 0.415 | 0.1026 | No |
| 7 | Retsat | 1547 | 0.410 | 0.1207 | No |
| 8 | Slc22a18 | 1726 | 0.389 | 0.1302 | No |
| 9 | Pex13 | 2078 | 0.356 | 0.1292 | No |
| 10 | Cyp7b1 | 2183 | 0.350 | 0.1407 | No |
| 11 | Hacl1 | 2581 | 0.318 | 0.1355 | No |
| 12 | Cat | 2758 | 0.304 | 0.1410 | No |
| 13 | Acsl5 | 3229 | 0.272 | 0.1299 | No |
| 14 | Slc27a2 | 3373 | 0.264 | 0.1351 | No |
| 15 | Amacr | 3659 | 0.249 | 0.1324 | No |
| 16 | Cyp27a1 | 4075 | 0.224 | 0.1218 | No |
| 17 | Hsd3b7 | 4118 | 0.224 | 0.1304 | No |
| 18 | Hsd17b4 | 4135 | 0.223 | 0.1402 | No |
| 19 | Rxra | 4147 | 0.222 | 0.1503 | No |
| 20 | Slc35b2 | 4165 | 0.221 | 0.1600 | No |
| 21 | Nudt12 | 4834 | 0.185 | 0.1345 | No |
| 22 | Gnpat | 5045 | 0.175 | 0.1321 | No |
| 23 | Idh1 | 5151 | 0.170 | 0.1349 | No |
| 24 | Nedd4 | 5179 | 0.169 | 0.1416 | No |
| 25 | Pex6 | 5575 | 0.152 | 0.1285 | No |
| 26 | Isoc1 | 5800 | 0.144 | 0.1239 | No |
| 27 | Pecr | 5967 | 0.136 | 0.1219 | No |
| 28 | Bcar3 | 5973 | 0.136 | 0.1282 | No |
| 29 | Slc23a2 | 5988 | 0.135 | 0.1339 | No |
| 30 | Abca3 | 6057 | 0.133 | 0.1368 | No |
| 31 | Cyp46a1 | 6200 | 0.126 | 0.1355 | No |
| 32 | Pex11a | 6554 | 0.111 | 0.1227 | No |
| 33 | Pex19 | 6611 | 0.109 | 0.1250 | No |
| 34 | Abca2 | 6821 | 0.100 | 0.1191 | No |
| 35 | Optn | 6824 | 0.100 | 0.1238 | No |
| 36 | Dhcr24 | 6831 | 0.100 | 0.1282 | No |
| 37 | Abca4 | 6839 | 0.100 | 0.1327 | No |
| 38 | Slc29a1 | 6942 | 0.096 | 0.1320 | No |
| 39 | Aldh1a1 | 7342 | 0.081 | 0.1154 | No |
| 40 | Idh2 | 7605 | 0.073 | 0.1054 | No |
| 41 | Abcd3 | 7701 | 0.070 | 0.1038 | No |
| 42 | Gclm | 8007 | 0.058 | 0.0909 | No |
| 43 | Abca9 | 9064 | 0.020 | 0.0375 | No |
| 44 | Nr3c2 | 9285 | 0.012 | 0.0267 | No |
| 45 | Abca8b | 9690 | 0.000 | 0.0059 | No |
| 46 | Pipox | 9693 | 0.000 | 0.0058 | No |
| 47 | Bmp6 | 9711 | 0.000 | 0.0049 | No |
| 48 | Pex7 | 9800 | -0.001 | 0.0005 | No |
| 49 | Rbp1 | 9905 | -0.005 | -0.0046 | No |
| 50 | Npc1 | 9914 | -0.005 | -0.0048 | No |
| 51 | Sod1 | 9954 | -0.007 | -0.0065 | No |
| 52 | Pex16 | 10053 | -0.010 | -0.0111 | No |
| 53 | Aqp9 | 10064 | -0.010 | -0.0111 | No |
| 54 | Sult2b1 | 10644 | -0.030 | -0.0395 | No |
| 55 | Aldh9a1 | 10735 | -0.034 | -0.0425 | No |
| 56 | Acsl1 | 10918 | -0.041 | -0.0499 | No |
| 57 | Klf1 | 11041 | -0.045 | -0.0540 | No |
| 58 | Abcd1 | 11181 | -0.050 | -0.0588 | No |
| 59 | Prdx5 | 11296 | -0.054 | -0.0621 | No |
| 60 | Lonp2 | 11361 | -0.056 | -0.0627 | No |
| 61 | Pex1 | 11447 | -0.059 | -0.0642 | No |
| 62 | Crot | 11938 | -0.074 | -0.0859 | No |
| 63 | Ar | 12396 | -0.090 | -0.1051 | No |
| 64 | Paox | 12551 | -0.095 | -0.1085 | No |
| 65 | Mlycd | 12585 | -0.096 | -0.1056 | No |
| 66 | Cyp39a1 | 12773 | -0.103 | -0.1103 | No |
| 67 | Lipe | 13007 | -0.112 | -0.1169 | No |
| 68 | Nr0b2 | 13083 | -0.115 | -0.1153 | No |
| 69 | Pex26 | 13417 | -0.127 | -0.1263 | No |
| 70 | Fads1 | 13500 | -0.131 | -0.1243 | No |
| 71 | Abcd2 | 13597 | -0.135 | -0.1227 | No |
| 72 | Pex11g | 13676 | -0.138 | -0.1201 | No |
| 73 | Pnpla8 | 13737 | -0.140 | -0.1165 | No |
| 74 | Abca5 | 13890 | -0.144 | -0.1174 | No |
| 75 | Pex12 | 13915 | -0.145 | -0.1117 | No |
| 76 | Hsd17b11 | 14067 | -0.151 | -0.1122 | No |
| 77 | Abca6 | 14369 | -0.163 | -0.1198 | No |
| 78 | Pfkm | 14702 | -0.178 | -0.1284 | No |
| 79 | Atxn1 | 14895 | -0.183 | -0.1295 | No |
| 80 | Soat2 | 16146 | -0.247 | -0.1820 | No |
| 81 | Idi1 | 16375 | -0.260 | -0.1813 | No |
| 82 | Slc23a1 | 17151 | -0.309 | -0.2064 | No |
| 83 | Phyh | 17788 | -0.364 | -0.2217 | No |
| 84 | Gstk1 | 17789 | -0.364 | -0.2042 | No |
| 85 | Pxmp2 | 18627 | -0.460 | -0.2253 | Yes |
| 86 | Nr1h4 | 18995 | -0.528 | -0.2188 | Yes |
| 87 | Dio2 | 19003 | -0.530 | -0.1937 | Yes |
| 88 | Efhc1 | 19148 | -0.579 | -0.1734 | Yes |
| 89 | Scp2 | 19228 | -0.615 | -0.1479 | Yes |
| 90 | Fads2 | 19406 | -0.742 | -0.1214 | Yes |
| 91 | Abca1 | 19435 | -0.782 | -0.0854 | Yes |
| 92 | Lck | 19460 | -0.817 | -0.0474 | Yes |
| 93 | Gnmt | 19504 | -1.042 | 0.0004 | Yes |