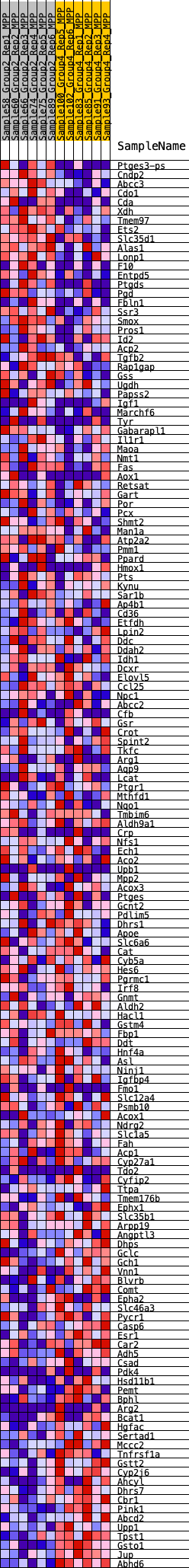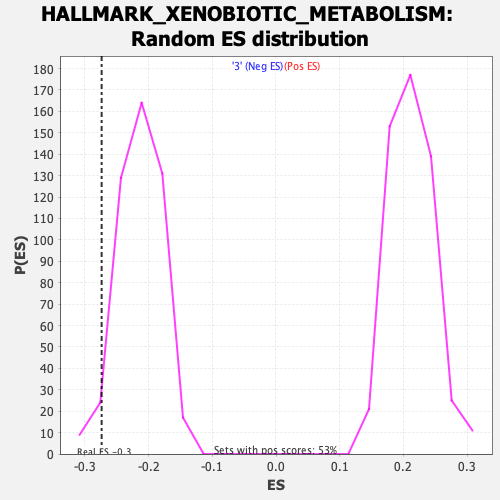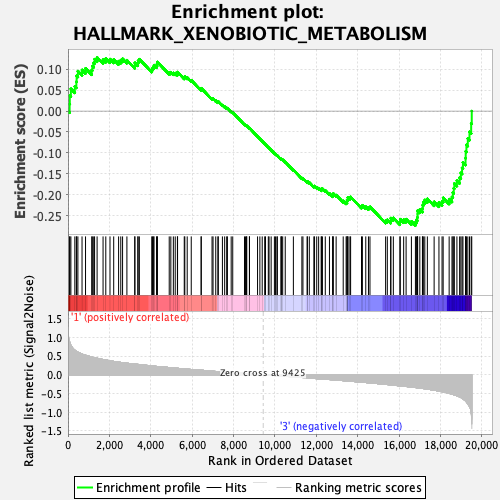
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.27378187 |
| Normalized Enrichment Score (NES) | -1.2752708 |
| Nominal p-value | 0.05907173 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.817 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ptges3-ps | 78 | 0.879 | 0.0169 | No |
| 2 | Cndp2 | 87 | 0.870 | 0.0371 | No |
| 3 | Abcc3 | 139 | 0.788 | 0.0533 | No |
| 4 | Cdo1 | 330 | 0.656 | 0.0590 | No |
| 5 | Cda | 403 | 0.627 | 0.0702 | No |
| 6 | Xdh | 418 | 0.621 | 0.0843 | No |
| 7 | Tmem97 | 478 | 0.603 | 0.0955 | No |
| 8 | Ets2 | 678 | 0.551 | 0.0984 | No |
| 9 | Slc35d1 | 847 | 0.523 | 0.1021 | No |
| 10 | Alas1 | 1146 | 0.475 | 0.0980 | No |
| 11 | Lonp1 | 1176 | 0.469 | 0.1077 | No |
| 12 | F10 | 1243 | 0.461 | 0.1153 | No |
| 13 | Entpd5 | 1284 | 0.456 | 0.1240 | No |
| 14 | Ptgds | 1406 | 0.443 | 0.1283 | No |
| 15 | Pgd | 1694 | 0.406 | 0.1232 | No |
| 16 | Fbln1 | 1826 | 0.395 | 0.1258 | No |
| 17 | Ssr3 | 2029 | 0.374 | 0.1242 | No |
| 18 | Smox | 2208 | 0.356 | 0.1235 | No |
| 19 | Pros1 | 2445 | 0.337 | 0.1193 | No |
| 20 | Id2 | 2550 | 0.330 | 0.1218 | No |
| 21 | Acp2 | 2637 | 0.324 | 0.1251 | No |
| 22 | Tgfb2 | 2846 | 0.308 | 0.1216 | No |
| 23 | Rap1gap | 3227 | 0.285 | 0.1088 | No |
| 24 | Gss | 3231 | 0.285 | 0.1154 | No |
| 25 | Ugdh | 3373 | 0.277 | 0.1147 | No |
| 26 | Papss2 | 3388 | 0.277 | 0.1206 | No |
| 27 | Igf1 | 3448 | 0.274 | 0.1240 | No |
| 28 | Marchf6 | 4047 | 0.237 | 0.0988 | No |
| 29 | Tyr | 4082 | 0.234 | 0.1026 | No |
| 30 | Gabarapl1 | 4126 | 0.232 | 0.1059 | No |
| 31 | Il1r1 | 4157 | 0.230 | 0.1098 | No |
| 32 | Maoa | 4279 | 0.223 | 0.1088 | No |
| 33 | Nmt1 | 4305 | 0.221 | 0.1128 | No |
| 34 | Fas | 4319 | 0.220 | 0.1174 | No |
| 35 | Aox1 | 4891 | 0.193 | 0.0925 | No |
| 36 | Retsat | 4969 | 0.189 | 0.0930 | No |
| 37 | Gart | 5090 | 0.184 | 0.0912 | No |
| 38 | Por | 5181 | 0.179 | 0.0908 | No |
| 39 | Pcx | 5287 | 0.173 | 0.0895 | No |
| 40 | Shmt2 | 5294 | 0.173 | 0.0933 | No |
| 41 | Man1a | 5617 | 0.157 | 0.0804 | No |
| 42 | Atp2a2 | 5644 | 0.156 | 0.0827 | No |
| 43 | Pmm1 | 5760 | 0.151 | 0.0804 | No |
| 44 | Ppard | 5955 | 0.141 | 0.0737 | No |
| 45 | Hmox1 | 6433 | 0.121 | 0.0519 | No |
| 46 | Pts | 6438 | 0.121 | 0.0546 | No |
| 47 | Kynu | 6960 | 0.096 | 0.0300 | No |
| 48 | Sar1b | 7008 | 0.094 | 0.0298 | No |
| 49 | Ap4b1 | 7139 | 0.088 | 0.0251 | No |
| 50 | Cd36 | 7243 | 0.084 | 0.0218 | No |
| 51 | Etfdh | 7262 | 0.083 | 0.0228 | No |
| 52 | Lpin2 | 7461 | 0.076 | 0.0144 | No |
| 53 | Ddc | 7570 | 0.072 | 0.0106 | No |
| 54 | Ddah2 | 7668 | 0.069 | 0.0072 | No |
| 55 | Idh1 | 7702 | 0.067 | 0.0071 | No |
| 56 | Dcxr | 7885 | 0.060 | -0.0009 | No |
| 57 | Elovl5 | 7960 | 0.056 | -0.0034 | No |
| 58 | Ccl25 | 8528 | 0.034 | -0.0319 | No |
| 59 | Npc1 | 8575 | 0.031 | -0.0335 | No |
| 60 | Abcc2 | 8628 | 0.029 | -0.0355 | No |
| 61 | Cfb | 8636 | 0.029 | -0.0351 | No |
| 62 | Gsr | 8765 | 0.024 | -0.0412 | No |
| 63 | Crot | 8770 | 0.024 | -0.0408 | No |
| 64 | Spint2 | 9159 | 0.010 | -0.0606 | No |
| 65 | Tkfc | 9271 | 0.006 | -0.0662 | No |
| 66 | Arg1 | 9384 | 0.002 | -0.0720 | No |
| 67 | Aqp9 | 9505 | -0.001 | -0.0782 | No |
| 68 | Lcat | 9543 | -0.002 | -0.0800 | No |
| 69 | Ptgr1 | 9679 | -0.008 | -0.0868 | No |
| 70 | Mthfd1 | 9714 | -0.009 | -0.0884 | No |
| 71 | Nqo1 | 9816 | -0.013 | -0.0933 | No |
| 72 | Tmbim6 | 9973 | -0.020 | -0.1009 | No |
| 73 | Aldh9a1 | 10024 | -0.022 | -0.1030 | No |
| 74 | Crp | 10095 | -0.024 | -0.1060 | No |
| 75 | Nfs1 | 10116 | -0.025 | -0.1065 | No |
| 76 | Ech1 | 10290 | -0.032 | -0.1146 | No |
| 77 | Aco2 | 10304 | -0.033 | -0.1145 | No |
| 78 | Upb1 | 10325 | -0.034 | -0.1147 | No |
| 79 | Mpp2 | 10374 | -0.036 | -0.1164 | No |
| 80 | Acox3 | 10497 | -0.041 | -0.1217 | No |
| 81 | Ptges | 10890 | -0.056 | -0.1406 | No |
| 82 | Gcnt2 | 11301 | -0.071 | -0.1601 | No |
| 83 | Pdlim5 | 11352 | -0.073 | -0.1610 | No |
| 84 | Dhrs1 | 11557 | -0.081 | -0.1696 | No |
| 85 | Apoe | 11565 | -0.082 | -0.1680 | No |
| 86 | Slc6a6 | 11660 | -0.087 | -0.1708 | No |
| 87 | Cat | 11880 | -0.097 | -0.1798 | No |
| 88 | Cyb5a | 11913 | -0.098 | -0.1791 | No |
| 89 | Hes6 | 12018 | -0.102 | -0.1821 | No |
| 90 | Pgrmc1 | 12111 | -0.107 | -0.1843 | No |
| 91 | Irf8 | 12238 | -0.112 | -0.1881 | No |
| 92 | Gnmt | 12242 | -0.112 | -0.1856 | No |
| 93 | Aldh2 | 12294 | -0.114 | -0.1856 | No |
| 94 | Hacl1 | 12432 | -0.119 | -0.1898 | No |
| 95 | Gstm4 | 12633 | -0.126 | -0.1972 | No |
| 96 | Fbp1 | 12787 | -0.132 | -0.2019 | No |
| 97 | Ddt | 12801 | -0.133 | -0.1994 | No |
| 98 | Hnf4a | 12817 | -0.133 | -0.1970 | No |
| 99 | Asl | 12956 | -0.139 | -0.2009 | No |
| 100 | Ninj1 | 13298 | -0.155 | -0.2148 | No |
| 101 | Igfbp4 | 13439 | -0.162 | -0.2182 | No |
| 102 | Fmo1 | 13482 | -0.164 | -0.2164 | No |
| 103 | Slc12a4 | 13484 | -0.164 | -0.2126 | No |
| 104 | Psmb10 | 13522 | -0.166 | -0.2105 | No |
| 105 | Acox1 | 13523 | -0.166 | -0.2066 | No |
| 106 | Ndrg2 | 13623 | -0.170 | -0.2077 | No |
| 107 | Slc1a5 | 13656 | -0.172 | -0.2052 | No |
| 108 | Fah | 14182 | -0.195 | -0.2277 | No |
| 109 | Acp1 | 14225 | -0.197 | -0.2252 | No |
| 110 | Cyp27a1 | 14388 | -0.205 | -0.2287 | No |
| 111 | Tdo2 | 14518 | -0.212 | -0.2303 | No |
| 112 | Cyfip2 | 14591 | -0.215 | -0.2289 | No |
| 113 | Ttpa | 15347 | -0.254 | -0.2618 | No |
| 114 | Tmem176b | 15429 | -0.259 | -0.2598 | No |
| 115 | Ephx1 | 15592 | -0.268 | -0.2618 | No |
| 116 | Slc35b1 | 15600 | -0.269 | -0.2558 | No |
| 117 | Arpp19 | 15720 | -0.276 | -0.2554 | No |
| 118 | Angptl3 | 16037 | -0.295 | -0.2647 | No |
| 119 | Dhps | 16054 | -0.296 | -0.2585 | No |
| 120 | Gclc | 16219 | -0.303 | -0.2597 | No |
| 121 | Gch1 | 16337 | -0.311 | -0.2584 | No |
| 122 | Vnn1 | 16593 | -0.326 | -0.2638 | No |
| 123 | Blvrb | 16787 | -0.339 | -0.2657 | Yes |
| 124 | Comt | 16835 | -0.343 | -0.2600 | Yes |
| 125 | Epha2 | 16882 | -0.347 | -0.2541 | Yes |
| 126 | Slc46a3 | 16891 | -0.347 | -0.2463 | Yes |
| 127 | Pycr1 | 16892 | -0.347 | -0.2380 | Yes |
| 128 | Casp6 | 17000 | -0.355 | -0.2351 | Yes |
| 129 | Esr1 | 17126 | -0.363 | -0.2329 | Yes |
| 130 | Car2 | 17138 | -0.364 | -0.2249 | Yes |
| 131 | Adh5 | 17168 | -0.366 | -0.2177 | Yes |
| 132 | Csad | 17240 | -0.372 | -0.2125 | Yes |
| 133 | Pdk4 | 17364 | -0.381 | -0.2098 | Yes |
| 134 | Hsd11b1 | 17691 | -0.410 | -0.2169 | Yes |
| 135 | Pemt | 17924 | -0.434 | -0.2185 | Yes |
| 136 | Bphl | 18074 | -0.450 | -0.2155 | Yes |
| 137 | Arg2 | 18127 | -0.456 | -0.2074 | Yes |
| 138 | Bcat1 | 18417 | -0.496 | -0.2105 | Yes |
| 139 | Hgfac | 18554 | -0.516 | -0.2053 | Yes |
| 140 | Sertad1 | 18596 | -0.523 | -0.1950 | Yes |
| 141 | Mccc2 | 18640 | -0.530 | -0.1846 | Yes |
| 142 | Tnfrsf1a | 18666 | -0.534 | -0.1732 | Yes |
| 143 | Gstt2 | 18792 | -0.559 | -0.1663 | Yes |
| 144 | Cyp2j6 | 18923 | -0.588 | -0.1591 | Yes |
| 145 | Ahcyl | 18982 | -0.605 | -0.1477 | Yes |
| 146 | Dhrs7 | 19042 | -0.628 | -0.1358 | Yes |
| 147 | Cbr1 | 19084 | -0.640 | -0.1227 | Yes |
| 148 | Pink1 | 19209 | -0.696 | -0.1126 | Yes |
| 149 | Abcd2 | 19217 | -0.701 | -0.0963 | Yes |
| 150 | Upp1 | 19252 | -0.727 | -0.0807 | Yes |
| 151 | Tpst1 | 19319 | -0.778 | -0.0657 | Yes |
| 152 | Gsto1 | 19400 | -0.848 | -0.0496 | Yes |
| 153 | Jup | 19481 | -1.018 | -0.0296 | Yes |
| 154 | Abhd6 | 19509 | -1.308 | 0.0002 | Yes |