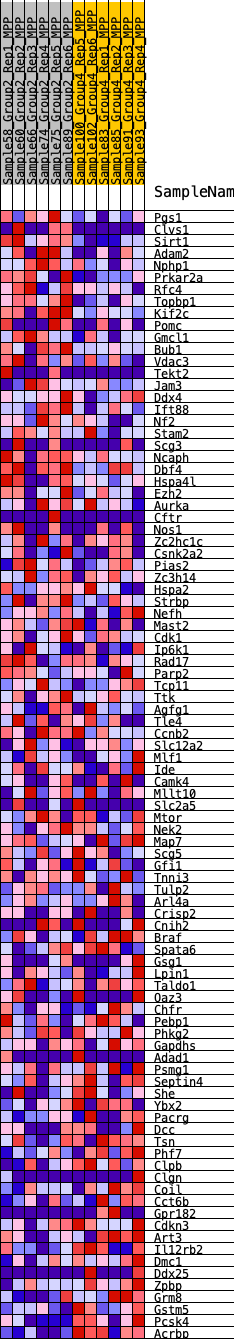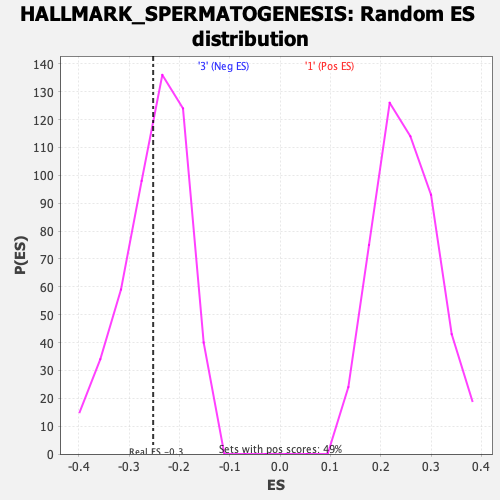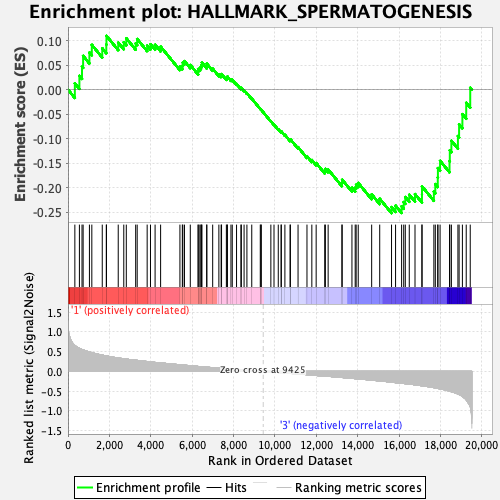
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.25224054 |
| Normalized Enrichment Score (NES) | -1.0136088 |
| Nominal p-value | 0.42094862 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pgs1 | 331 | 0.655 | 0.0129 | No |
| 2 | Clvs1 | 551 | 0.582 | 0.0282 | No |
| 3 | Sirt1 | 677 | 0.551 | 0.0470 | No |
| 4 | Adam2 | 731 | 0.541 | 0.0689 | No |
| 5 | Nphp1 | 1035 | 0.491 | 0.0758 | No |
| 6 | Prkar2a | 1151 | 0.474 | 0.0915 | No |
| 7 | Rfc4 | 1655 | 0.411 | 0.0844 | No |
| 8 | Topbp1 | 1851 | 0.393 | 0.0923 | No |
| 9 | Kif2c | 1859 | 0.392 | 0.1098 | No |
| 10 | Pomc | 2427 | 0.339 | 0.0961 | No |
| 11 | Gmcl1 | 2695 | 0.319 | 0.0969 | No |
| 12 | Bub1 | 2817 | 0.310 | 0.1048 | No |
| 13 | Vdac3 | 3270 | 0.282 | 0.0944 | No |
| 14 | Tekt2 | 3343 | 0.278 | 0.1034 | No |
| 15 | Jam3 | 3825 | 0.250 | 0.0901 | No |
| 16 | Ddx4 | 3986 | 0.240 | 0.0928 | No |
| 17 | Ift88 | 4208 | 0.227 | 0.0917 | No |
| 18 | Nf2 | 4475 | 0.215 | 0.0878 | No |
| 19 | Stam2 | 5408 | 0.167 | 0.0475 | No |
| 20 | Scg3 | 5525 | 0.161 | 0.0489 | No |
| 21 | Ncaph | 5548 | 0.161 | 0.0551 | No |
| 22 | Dbf4 | 5627 | 0.157 | 0.0582 | No |
| 23 | Hspa4l | 5909 | 0.143 | 0.0503 | No |
| 24 | Ezh2 | 6289 | 0.125 | 0.0365 | No |
| 25 | Aurka | 6290 | 0.125 | 0.0422 | No |
| 26 | Cftr | 6371 | 0.122 | 0.0437 | No |
| 27 | Nos1 | 6420 | 0.121 | 0.0468 | No |
| 28 | Zc2hc1c | 6453 | 0.120 | 0.0506 | No |
| 29 | Csnk2a2 | 6471 | 0.119 | 0.0551 | No |
| 30 | Pias2 | 6704 | 0.108 | 0.0481 | No |
| 31 | Zc3h14 | 6706 | 0.108 | 0.0530 | No |
| 32 | Hspa2 | 6991 | 0.095 | 0.0427 | No |
| 33 | Strbp | 7290 | 0.082 | 0.0311 | No |
| 34 | Nefh | 7399 | 0.079 | 0.0292 | No |
| 35 | Mast2 | 7419 | 0.078 | 0.0318 | No |
| 36 | Cdk1 | 7652 | 0.070 | 0.0230 | No |
| 37 | Ip6k1 | 7689 | 0.068 | 0.0242 | No |
| 38 | Rad17 | 7701 | 0.067 | 0.0268 | No |
| 39 | Parp2 | 7874 | 0.060 | 0.0207 | No |
| 40 | Tcp11 | 7937 | 0.057 | 0.0201 | No |
| 41 | Ttk | 8144 | 0.049 | 0.0117 | No |
| 42 | Agfg1 | 8355 | 0.041 | 0.0028 | No |
| 43 | Tle4 | 8376 | 0.040 | 0.0036 | No |
| 44 | Ccnb2 | 8508 | 0.035 | -0.0015 | No |
| 45 | Slc12a2 | 8645 | 0.029 | -0.0072 | No |
| 46 | Mlf1 | 8879 | 0.019 | -0.0184 | No |
| 47 | Ide | 9291 | 0.005 | -0.0393 | No |
| 48 | Camk4 | 9323 | 0.004 | -0.0407 | No |
| 49 | Mllt10 | 9338 | 0.003 | -0.0413 | No |
| 50 | Slc2a5 | 9802 | -0.012 | -0.0646 | No |
| 51 | Mtor | 9954 | -0.019 | -0.0715 | No |
| 52 | Nek2 | 10160 | -0.027 | -0.0808 | No |
| 53 | Map7 | 10297 | -0.033 | -0.0863 | No |
| 54 | Scg5 | 10307 | -0.033 | -0.0853 | No |
| 55 | Gfi1 | 10480 | -0.040 | -0.0923 | No |
| 56 | Tnni3 | 10731 | -0.048 | -0.1030 | No |
| 57 | Tulp2 | 10746 | -0.049 | -0.1015 | No |
| 58 | Arl4a | 11116 | -0.064 | -0.1175 | No |
| 59 | Crisp2 | 11547 | -0.081 | -0.1360 | No |
| 60 | Cnih2 | 11782 | -0.092 | -0.1438 | No |
| 61 | Braf | 11991 | -0.101 | -0.1499 | No |
| 62 | Spata6 | 12405 | -0.118 | -0.1658 | No |
| 63 | Gsg1 | 12436 | -0.120 | -0.1619 | No |
| 64 | Lpin1 | 12570 | -0.124 | -0.1631 | No |
| 65 | Taldo1 | 13235 | -0.152 | -0.1903 | No |
| 66 | Oaz3 | 13247 | -0.152 | -0.1839 | No |
| 67 | Chfr | 13720 | -0.174 | -0.2003 | No |
| 68 | Pebp1 | 13877 | -0.181 | -0.2001 | No |
| 69 | Phkg2 | 13923 | -0.183 | -0.1940 | No |
| 70 | Gapdhs | 14024 | -0.188 | -0.1906 | No |
| 71 | Adad1 | 14673 | -0.220 | -0.2139 | No |
| 72 | Psmg1 | 15062 | -0.240 | -0.2229 | No |
| 73 | Septin4 | 15633 | -0.271 | -0.2399 | Yes |
| 74 | She | 15828 | -0.283 | -0.2369 | Yes |
| 75 | Ybx2 | 16120 | -0.300 | -0.2382 | Yes |
| 76 | Pacrg | 16222 | -0.303 | -0.2296 | Yes |
| 77 | Dcc | 16298 | -0.309 | -0.2193 | Yes |
| 78 | Tsn | 16490 | -0.320 | -0.2146 | Yes |
| 79 | Phf7 | 16767 | -0.337 | -0.2134 | Yes |
| 80 | Clpb | 17103 | -0.362 | -0.2141 | Yes |
| 81 | Clgn | 17105 | -0.362 | -0.1976 | Yes |
| 82 | Coil | 17679 | -0.409 | -0.2084 | Yes |
| 83 | Cct6b | 17748 | -0.415 | -0.1929 | Yes |
| 84 | Gpr182 | 17867 | -0.428 | -0.1794 | Yes |
| 85 | Cdkn3 | 17874 | -0.429 | -0.1602 | Yes |
| 86 | Art3 | 17979 | -0.440 | -0.1454 | Yes |
| 87 | Il12rb2 | 18435 | -0.498 | -0.1461 | Yes |
| 88 | Dmc1 | 18449 | -0.500 | -0.1239 | Yes |
| 89 | Ddx25 | 18525 | -0.511 | -0.1044 | Yes |
| 90 | Zpbp | 18841 | -0.569 | -0.0946 | Yes |
| 91 | Grm8 | 18898 | -0.584 | -0.0708 | Yes |
| 92 | Gstm5 | 19057 | -0.632 | -0.0501 | Yes |
| 93 | Pcsk4 | 19242 | -0.717 | -0.0268 | Yes |
| 94 | Acrbp | 19436 | -0.890 | 0.0039 | Yes |