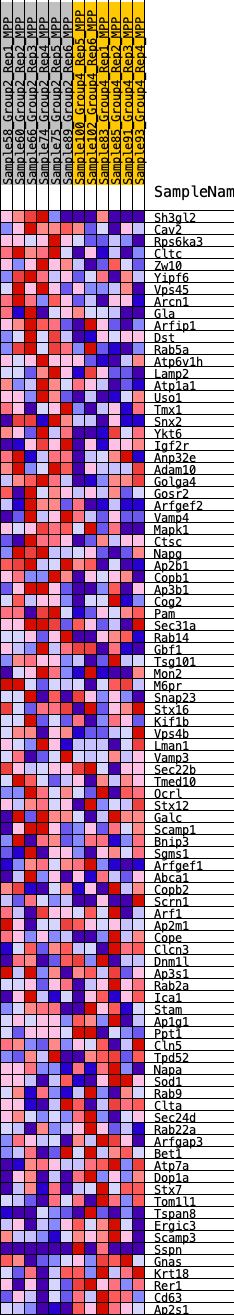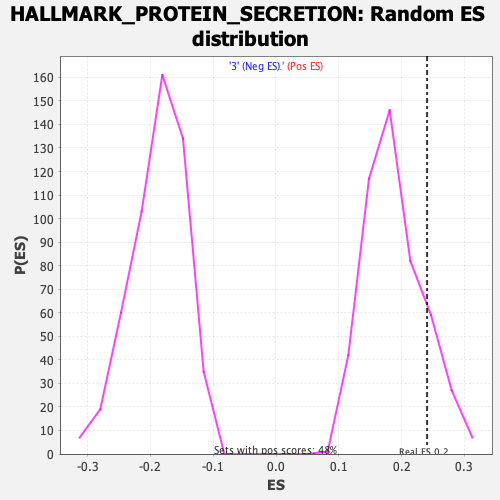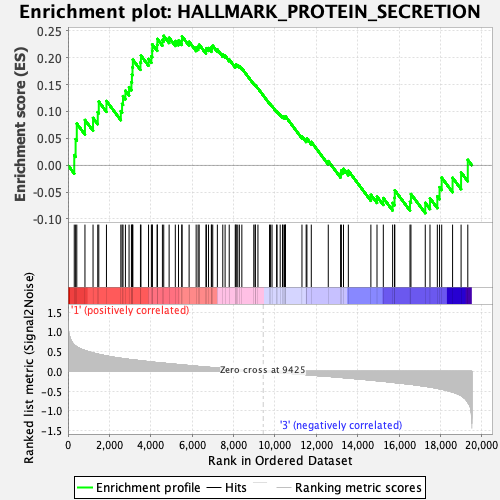
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.24067774 |
| Normalized Enrichment Score (NES) | 1.2742234 |
| Nominal p-value | 0.14345114 |
| FDR q-value | 0.60619545 |
| FWER p-Value | 0.826 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sh3gl2 | 302 | 0.669 | 0.0188 | Yes |
| 2 | Cav2 | 368 | 0.641 | 0.0484 | Yes |
| 3 | Rps6ka3 | 425 | 0.619 | 0.0773 | Yes |
| 4 | Cltc | 819 | 0.527 | 0.0842 | Yes |
| 5 | Zw10 | 1210 | 0.465 | 0.0880 | Yes |
| 6 | Yipf6 | 1438 | 0.437 | 0.0988 | Yes |
| 7 | Vps45 | 1488 | 0.430 | 0.1183 | Yes |
| 8 | Arcn1 | 1858 | 0.392 | 0.1195 | Yes |
| 9 | Gla | 2553 | 0.329 | 0.1007 | Yes |
| 10 | Arfip1 | 2613 | 0.325 | 0.1143 | Yes |
| 11 | Dst | 2658 | 0.322 | 0.1286 | Yes |
| 12 | Rab5a | 2774 | 0.313 | 0.1387 | Yes |
| 13 | Atp6v1h | 2950 | 0.302 | 0.1452 | Yes |
| 14 | Lamp2 | 3065 | 0.296 | 0.1546 | Yes |
| 15 | Atp1a1 | 3089 | 0.295 | 0.1685 | Yes |
| 16 | Uso1 | 3117 | 0.293 | 0.1822 | Yes |
| 17 | Tmx1 | 3134 | 0.291 | 0.1963 | Yes |
| 18 | Snx2 | 3501 | 0.270 | 0.1913 | Yes |
| 19 | Ykt6 | 3525 | 0.268 | 0.2039 | Yes |
| 20 | Igf2r | 3891 | 0.245 | 0.1977 | Yes |
| 21 | Anp32e | 4029 | 0.238 | 0.2029 | Yes |
| 22 | Adam10 | 4067 | 0.235 | 0.2130 | Yes |
| 23 | Golga4 | 4073 | 0.235 | 0.2248 | Yes |
| 24 | Gosr2 | 4308 | 0.221 | 0.2241 | Yes |
| 25 | Arfgef2 | 4318 | 0.220 | 0.2350 | Yes |
| 26 | Vamp4 | 4562 | 0.210 | 0.2333 | Yes |
| 27 | Mapk1 | 4626 | 0.207 | 0.2407 | Yes |
| 28 | Ctsc | 4886 | 0.193 | 0.2373 | No |
| 29 | Napg | 5185 | 0.179 | 0.2311 | No |
| 30 | Ap2b1 | 5339 | 0.170 | 0.2320 | No |
| 31 | Copb1 | 5499 | 0.162 | 0.2321 | No |
| 32 | Ap3b1 | 5509 | 0.162 | 0.2400 | No |
| 33 | Cog2 | 5852 | 0.146 | 0.2298 | No |
| 34 | Pam | 6192 | 0.130 | 0.2191 | No |
| 35 | Sec31a | 6285 | 0.125 | 0.2208 | No |
| 36 | Rab14 | 6338 | 0.123 | 0.2244 | No |
| 37 | Gbf1 | 6665 | 0.111 | 0.2133 | No |
| 38 | Tsg101 | 6689 | 0.109 | 0.2177 | No |
| 39 | Mon2 | 6788 | 0.104 | 0.2180 | No |
| 40 | M6pr | 6926 | 0.097 | 0.2159 | No |
| 41 | Snap23 | 6943 | 0.096 | 0.2201 | No |
| 42 | Stx16 | 6994 | 0.094 | 0.2223 | No |
| 43 | Kif1b | 7217 | 0.085 | 0.2153 | No |
| 44 | Vps4b | 7479 | 0.076 | 0.2057 | No |
| 45 | Lman1 | 7591 | 0.072 | 0.2037 | No |
| 46 | Vamp3 | 7790 | 0.065 | 0.1968 | No |
| 47 | Sec22b | 8082 | 0.052 | 0.1845 | No |
| 48 | Tmed10 | 8117 | 0.051 | 0.1853 | No |
| 49 | Ocrl | 8124 | 0.050 | 0.1876 | No |
| 50 | Stx12 | 8202 | 0.047 | 0.1860 | No |
| 51 | Galc | 8280 | 0.044 | 0.1843 | No |
| 52 | Scamp1 | 8400 | 0.039 | 0.1802 | No |
| 53 | Bnip3 | 8977 | 0.016 | 0.1513 | No |
| 54 | Sgms1 | 9044 | 0.014 | 0.1487 | No |
| 55 | Arfgef1 | 9050 | 0.014 | 0.1491 | No |
| 56 | Abca1 | 9178 | 0.009 | 0.1430 | No |
| 57 | Copb2 | 9738 | -0.010 | 0.1148 | No |
| 58 | Scrn1 | 9789 | -0.011 | 0.1128 | No |
| 59 | Arf1 | 9859 | -0.015 | 0.1100 | No |
| 60 | Ap2m1 | 10081 | -0.023 | 0.0998 | No |
| 61 | Cope | 10100 | -0.024 | 0.1001 | No |
| 62 | Clcn3 | 10253 | -0.031 | 0.0938 | No |
| 63 | Dnm1l | 10365 | -0.035 | 0.0899 | No |
| 64 | Ap3s1 | 10391 | -0.037 | 0.0905 | No |
| 65 | Rab2a | 10460 | -0.039 | 0.0891 | No |
| 66 | Ica1 | 10504 | -0.041 | 0.0889 | No |
| 67 | Stam | 10505 | -0.041 | 0.0910 | No |
| 68 | Ap1g1 | 11305 | -0.071 | 0.0536 | No |
| 69 | Ppt1 | 11508 | -0.079 | 0.0472 | No |
| 70 | Cln5 | 11544 | -0.081 | 0.0496 | No |
| 71 | Tpd52 | 11756 | -0.091 | 0.0434 | No |
| 72 | Napa | 12576 | -0.124 | 0.0076 | No |
| 73 | Sod1 | 13171 | -0.149 | -0.0153 | No |
| 74 | Rab9 | 13210 | -0.150 | -0.0096 | No |
| 75 | Clta | 13312 | -0.156 | -0.0068 | No |
| 76 | Sec24d | 13546 | -0.167 | -0.0102 | No |
| 77 | Rab22a | 14635 | -0.218 | -0.0551 | No |
| 78 | Arfgap3 | 14931 | -0.233 | -0.0583 | No |
| 79 | Bet1 | 15236 | -0.249 | -0.0611 | No |
| 80 | Atp7a | 15686 | -0.274 | -0.0702 | No |
| 81 | Dop1a | 15777 | -0.280 | -0.0605 | No |
| 82 | Stx7 | 15791 | -0.280 | -0.0467 | No |
| 83 | Tom1l1 | 16526 | -0.321 | -0.0680 | No |
| 84 | Tspan8 | 16571 | -0.324 | -0.0536 | No |
| 85 | Ergic3 | 17263 | -0.373 | -0.0700 | No |
| 86 | Scamp3 | 17491 | -0.392 | -0.0616 | No |
| 87 | Sspn | 17846 | -0.426 | -0.0579 | No |
| 88 | Gnas | 17953 | -0.437 | -0.0409 | No |
| 89 | Krt18 | 18057 | -0.448 | -0.0232 | No |
| 90 | Rer1 | 18583 | -0.521 | -0.0235 | No |
| 91 | Cd63 | 18994 | -0.609 | -0.0133 | No |
| 92 | Ap2s1 | 19318 | -0.777 | 0.0100 | No |