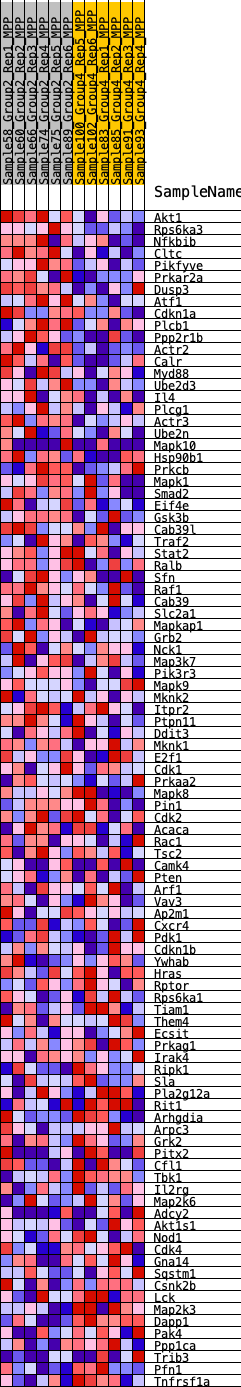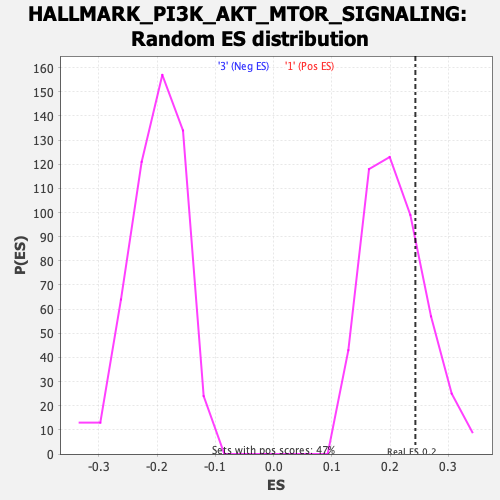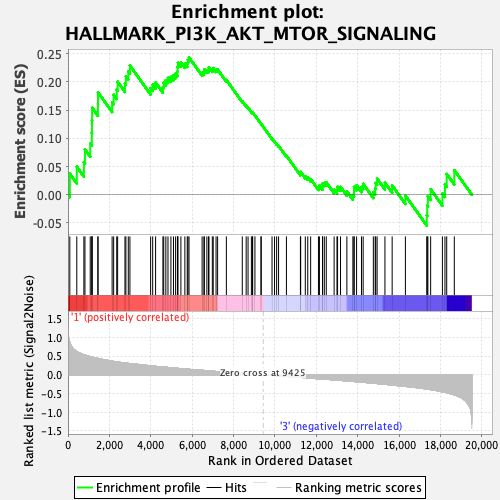
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.24352376 |
| Normalized Enrichment Score (NES) | 1.1682891 |
| Nominal p-value | 0.23839663 |
| FDR q-value | 0.8852899 |
| FWER p-Value | 0.943 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Akt1 | 88 | 0.870 | 0.0372 | Yes |
| 2 | Rps6ka3 | 425 | 0.619 | 0.0497 | Yes |
| 3 | Nfkbib | 767 | 0.535 | 0.0578 | Yes |
| 4 | Cltc | 819 | 0.527 | 0.0804 | Yes |
| 5 | Pikfyve | 1074 | 0.486 | 0.0907 | Yes |
| 6 | Prkar2a | 1151 | 0.474 | 0.1095 | Yes |
| 7 | Dusp3 | 1158 | 0.473 | 0.1319 | Yes |
| 8 | Atf1 | 1166 | 0.472 | 0.1542 | Yes |
| 9 | Cdkn1a | 1446 | 0.437 | 0.1608 | Yes |
| 10 | Plcb1 | 1449 | 0.437 | 0.1817 | Yes |
| 11 | Ppp2r1b | 2132 | 0.362 | 0.1639 | Yes |
| 12 | Actr2 | 2206 | 0.357 | 0.1773 | Yes |
| 13 | Calr | 2355 | 0.344 | 0.1862 | Yes |
| 14 | Myd88 | 2398 | 0.341 | 0.2004 | Yes |
| 15 | Ube2d3 | 2752 | 0.314 | 0.1973 | Yes |
| 16 | Il4 | 2797 | 0.311 | 0.2100 | Yes |
| 17 | Plcg1 | 2913 | 0.304 | 0.2187 | Yes |
| 18 | Actr3 | 2992 | 0.300 | 0.2291 | Yes |
| 19 | Ube2n | 3992 | 0.239 | 0.1891 | Yes |
| 20 | Mapk10 | 4092 | 0.234 | 0.1952 | Yes |
| 21 | Hsp90b1 | 4232 | 0.225 | 0.1989 | Yes |
| 22 | Prkcb | 4586 | 0.209 | 0.1907 | Yes |
| 23 | Mapk1 | 4626 | 0.207 | 0.1987 | Yes |
| 24 | Smad2 | 4728 | 0.202 | 0.2032 | Yes |
| 25 | Eif4e | 4831 | 0.196 | 0.2073 | Yes |
| 26 | Gsk3b | 4973 | 0.189 | 0.2091 | Yes |
| 27 | Cab39l | 5096 | 0.184 | 0.2117 | Yes |
| 28 | Traf2 | 5200 | 0.178 | 0.2149 | Yes |
| 29 | Stat2 | 5289 | 0.173 | 0.2187 | Yes |
| 30 | Ralb | 5293 | 0.173 | 0.2268 | Yes |
| 31 | Sfn | 5314 | 0.171 | 0.2340 | Yes |
| 32 | Raf1 | 5455 | 0.165 | 0.2347 | Yes |
| 33 | Cab39 | 5647 | 0.156 | 0.2323 | Yes |
| 34 | Slc2a1 | 5762 | 0.151 | 0.2337 | Yes |
| 35 | Mapkap1 | 5795 | 0.149 | 0.2392 | Yes |
| 36 | Grb2 | 5848 | 0.146 | 0.2435 | Yes |
| 37 | Nck1 | 6480 | 0.119 | 0.2167 | No |
| 38 | Map3k7 | 6567 | 0.115 | 0.2178 | No |
| 39 | Pik3r3 | 6589 | 0.114 | 0.2222 | No |
| 40 | Mapk9 | 6710 | 0.108 | 0.2212 | No |
| 41 | Mknk2 | 6792 | 0.103 | 0.2220 | No |
| 42 | Itpr2 | 6810 | 0.103 | 0.2261 | No |
| 43 | Ptpn11 | 6973 | 0.095 | 0.2223 | No |
| 44 | Ddit3 | 7017 | 0.093 | 0.2246 | No |
| 45 | Mknk1 | 7154 | 0.088 | 0.2218 | No |
| 46 | E2f1 | 7229 | 0.084 | 0.2220 | No |
| 47 | Cdk1 | 7652 | 0.070 | 0.2036 | No |
| 48 | Prkaa2 | 8421 | 0.038 | 0.1659 | No |
| 49 | Mapk8 | 8609 | 0.030 | 0.1577 | No |
| 50 | Pin1 | 8700 | 0.026 | 0.1543 | No |
| 51 | Cdk2 | 8861 | 0.020 | 0.1470 | No |
| 52 | Acaca | 8917 | 0.017 | 0.1450 | No |
| 53 | Rac1 | 8921 | 0.017 | 0.1457 | No |
| 54 | Tsc2 | 9032 | 0.014 | 0.1407 | No |
| 55 | Camk4 | 9323 | 0.004 | 0.1260 | No |
| 56 | Pten | 9337 | 0.003 | 0.1254 | No |
| 57 | Arf1 | 9859 | -0.015 | 0.0993 | No |
| 58 | Vav3 | 9980 | -0.020 | 0.0941 | No |
| 59 | Ap2m1 | 10081 | -0.023 | 0.0901 | No |
| 60 | Cxcr4 | 10175 | -0.027 | 0.0866 | No |
| 61 | Pdk1 | 10554 | -0.042 | 0.0691 | No |
| 62 | Cdkn1b | 11230 | -0.069 | 0.0377 | No |
| 63 | Ywhab | 11243 | -0.069 | 0.0404 | No |
| 64 | Hras | 11466 | -0.078 | 0.0327 | No |
| 65 | Rptor | 11585 | -0.083 | 0.0306 | No |
| 66 | Rps6ka1 | 11725 | -0.089 | 0.0277 | No |
| 67 | Tiam1 | 12102 | -0.106 | 0.0135 | No |
| 68 | Them4 | 12149 | -0.108 | 0.0163 | No |
| 69 | Ecsit | 12299 | -0.114 | 0.0141 | No |
| 70 | Prkag1 | 12301 | -0.114 | 0.0195 | No |
| 71 | Irak4 | 12385 | -0.118 | 0.0209 | No |
| 72 | Ripk1 | 12479 | -0.121 | 0.0219 | No |
| 73 | Sla | 12859 | -0.135 | 0.0089 | No |
| 74 | Pla2g12a | 12997 | -0.141 | 0.0086 | No |
| 75 | Rit1 | 13026 | -0.142 | 0.0140 | No |
| 76 | Arhgdia | 13168 | -0.148 | 0.0138 | No |
| 77 | Arpc3 | 13475 | -0.164 | 0.0059 | No |
| 78 | Grk2 | 13766 | -0.177 | -0.0005 | No |
| 79 | Pitx2 | 13822 | -0.179 | 0.0053 | No |
| 80 | Cfl1 | 13823 | -0.179 | 0.0139 | No |
| 81 | Tbk1 | 13944 | -0.184 | 0.0166 | No |
| 82 | Il2rg | 14181 | -0.195 | 0.0138 | No |
| 83 | Map2k6 | 14262 | -0.200 | 0.0192 | No |
| 84 | Adcy2 | 14754 | -0.222 | 0.0046 | No |
| 85 | Akt1s1 | 14843 | -0.228 | 0.0110 | No |
| 86 | Nod1 | 14868 | -0.229 | 0.0208 | No |
| 87 | Cdk4 | 14936 | -0.233 | 0.0285 | No |
| 88 | Gna14 | 15314 | -0.253 | 0.0213 | No |
| 89 | Sqstm1 | 15663 | -0.272 | 0.0164 | No |
| 90 | Csnk2b | 16303 | -0.309 | -0.0017 | No |
| 91 | Lck | 17337 | -0.379 | -0.0367 | No |
| 92 | Map2k3 | 17358 | -0.381 | -0.0194 | No |
| 93 | Dapp1 | 17389 | -0.384 | -0.0025 | No |
| 94 | Pak4 | 17523 | -0.395 | 0.0096 | No |
| 95 | Ppp1ca | 18091 | -0.452 | 0.0021 | No |
| 96 | Trib3 | 18214 | -0.466 | 0.0182 | No |
| 97 | Pfn1 | 18299 | -0.477 | 0.0368 | No |
| 98 | Tnfrsf1a | 18666 | -0.534 | 0.0436 | No |