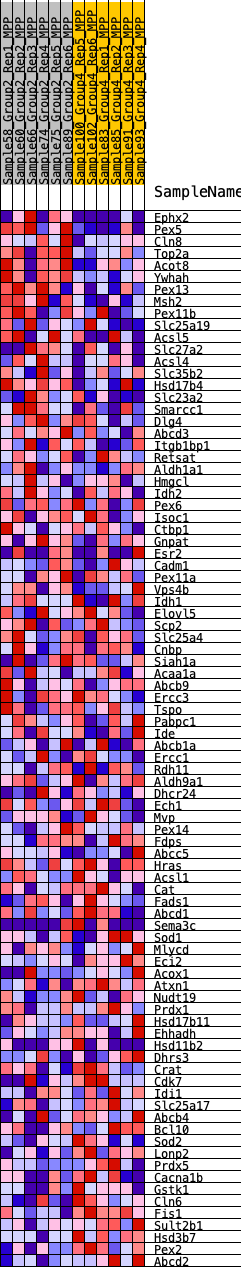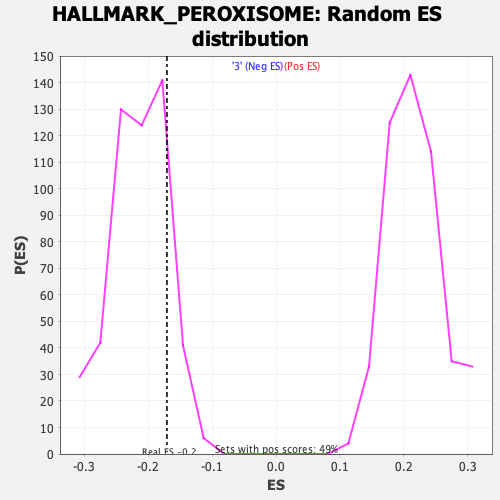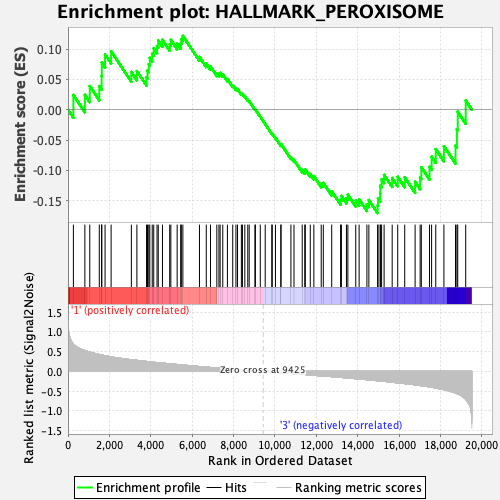
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.17045976 |
| Normalized Enrichment Score (NES) | -0.7972191 |
| Nominal p-value | 0.8440546 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ephx2 | 260 | 0.691 | 0.0243 | No |
| 2 | Pex5 | 815 | 0.528 | 0.0246 | No |
| 3 | Cln8 | 1052 | 0.488 | 0.0390 | No |
| 4 | Top2a | 1512 | 0.428 | 0.0387 | No |
| 5 | Acot8 | 1624 | 0.414 | 0.0556 | No |
| 6 | Ywhah | 1633 | 0.413 | 0.0777 | No |
| 7 | Pex13 | 1791 | 0.398 | 0.0913 | No |
| 8 | Msh2 | 2083 | 0.368 | 0.0964 | No |
| 9 | Pex11b | 3061 | 0.296 | 0.0623 | No |
| 10 | Slc25a19 | 3329 | 0.278 | 0.0637 | No |
| 11 | Acsl5 | 3795 | 0.251 | 0.0535 | No |
| 12 | Slc27a2 | 3841 | 0.249 | 0.0647 | No |
| 13 | Acsl4 | 3903 | 0.244 | 0.0749 | No |
| 14 | Slc35b2 | 3953 | 0.241 | 0.0855 | No |
| 15 | Hsd17b4 | 4070 | 0.235 | 0.0924 | No |
| 16 | Slc23a2 | 4143 | 0.231 | 0.1012 | No |
| 17 | Smarcc1 | 4302 | 0.221 | 0.1052 | No |
| 18 | Dlg4 | 4361 | 0.219 | 0.1141 | No |
| 19 | Abcd3 | 4564 | 0.210 | 0.1152 | No |
| 20 | Itgb1bp1 | 4914 | 0.192 | 0.1077 | No |
| 21 | Retsat | 4969 | 0.189 | 0.1152 | No |
| 22 | Aldh1a1 | 5274 | 0.174 | 0.1090 | No |
| 23 | Hmgcl | 5445 | 0.165 | 0.1093 | No |
| 24 | Idh2 | 5476 | 0.164 | 0.1167 | No |
| 25 | Pex6 | 5550 | 0.161 | 0.1217 | No |
| 26 | Isoc1 | 6352 | 0.122 | 0.0871 | No |
| 27 | Ctbp1 | 6681 | 0.109 | 0.0762 | No |
| 28 | Gnpat | 6885 | 0.099 | 0.0711 | No |
| 29 | Esr2 | 7192 | 0.086 | 0.0601 | No |
| 30 | Cadm1 | 7294 | 0.082 | 0.0593 | No |
| 31 | Pex11a | 7351 | 0.080 | 0.0608 | No |
| 32 | Vps4b | 7479 | 0.076 | 0.0584 | No |
| 33 | Idh1 | 7702 | 0.067 | 0.0506 | No |
| 34 | Elovl5 | 7960 | 0.056 | 0.0405 | No |
| 35 | Scp2 | 8115 | 0.051 | 0.0353 | No |
| 36 | Slc25a4 | 8192 | 0.047 | 0.0340 | No |
| 37 | Cnbp | 8377 | 0.040 | 0.0267 | No |
| 38 | Siah1a | 8429 | 0.038 | 0.0261 | No |
| 39 | Acaa1a | 8544 | 0.033 | 0.0221 | No |
| 40 | Abcb9 | 8687 | 0.027 | 0.0162 | No |
| 41 | Ercc3 | 8755 | 0.024 | 0.0141 | No |
| 42 | Tspo | 9033 | 0.014 | 0.0006 | No |
| 43 | Pabpc1 | 9055 | 0.013 | 0.0002 | No |
| 44 | Ide | 9291 | 0.005 | -0.0116 | No |
| 45 | Abcb1a | 9529 | -0.001 | -0.0237 | No |
| 46 | Ercc1 | 9848 | -0.014 | -0.0393 | No |
| 47 | Rdh11 | 9868 | -0.015 | -0.0394 | No |
| 48 | Aldh9a1 | 10024 | -0.022 | -0.0462 | No |
| 49 | Dhcr24 | 10287 | -0.032 | -0.0580 | No |
| 50 | Ech1 | 10290 | -0.032 | -0.0564 | No |
| 51 | Mvp | 10771 | -0.051 | -0.0783 | No |
| 52 | Pex14 | 10920 | -0.057 | -0.0828 | No |
| 53 | Fdps | 11317 | -0.072 | -0.0993 | No |
| 54 | Abcc5 | 11441 | -0.077 | -0.1015 | No |
| 55 | Hras | 11466 | -0.078 | -0.0985 | No |
| 56 | Acsl1 | 11707 | -0.089 | -0.1060 | No |
| 57 | Cat | 11880 | -0.097 | -0.1096 | No |
| 58 | Fads1 | 12235 | -0.111 | -0.1217 | No |
| 59 | Abcd1 | 12336 | -0.116 | -0.1206 | No |
| 60 | Sema3c | 12742 | -0.130 | -0.1343 | No |
| 61 | Sod1 | 13171 | -0.149 | -0.1482 | No |
| 62 | Mlycd | 13211 | -0.151 | -0.1420 | No |
| 63 | Eci2 | 13454 | -0.163 | -0.1456 | No |
| 64 | Acox1 | 13523 | -0.166 | -0.1401 | No |
| 65 | Atxn1 | 13904 | -0.183 | -0.1497 | No |
| 66 | Nudt19 | 14067 | -0.190 | -0.1476 | No |
| 67 | Prdx1 | 14443 | -0.208 | -0.1556 | No |
| 68 | Hsd17b11 | 14538 | -0.213 | -0.1488 | No |
| 69 | Ehhadh | 14960 | -0.235 | -0.1577 | Yes |
| 70 | Hsd11b2 | 14982 | -0.236 | -0.1459 | Yes |
| 71 | Dhrs3 | 15086 | -0.242 | -0.1380 | Yes |
| 72 | Crat | 15090 | -0.242 | -0.1250 | Yes |
| 73 | Cdk7 | 15153 | -0.244 | -0.1149 | Yes |
| 74 | Idi1 | 15274 | -0.251 | -0.1074 | Yes |
| 75 | Slc25a17 | 15664 | -0.272 | -0.1126 | Yes |
| 76 | Abcb4 | 15932 | -0.289 | -0.1106 | Yes |
| 77 | Bcl10 | 16270 | -0.307 | -0.1112 | Yes |
| 78 | Sod2 | 16772 | -0.338 | -0.1186 | Yes |
| 79 | Lonp2 | 17015 | -0.356 | -0.1117 | Yes |
| 80 | Prdx5 | 17072 | -0.360 | -0.0949 | Yes |
| 81 | Cacna1b | 17468 | -0.390 | -0.0940 | Yes |
| 82 | Gstk1 | 17569 | -0.400 | -0.0773 | Yes |
| 83 | Cln6 | 17770 | -0.417 | -0.0649 | Yes |
| 84 | Fis1 | 18164 | -0.459 | -0.0601 | Yes |
| 85 | Sult2b1 | 18724 | -0.545 | -0.0592 | Yes |
| 86 | Hsd3b7 | 18791 | -0.559 | -0.0321 | Yes |
| 87 | Pex2 | 18832 | -0.567 | -0.0033 | Yes |
| 88 | Abcd2 | 19217 | -0.701 | 0.0152 | Yes |