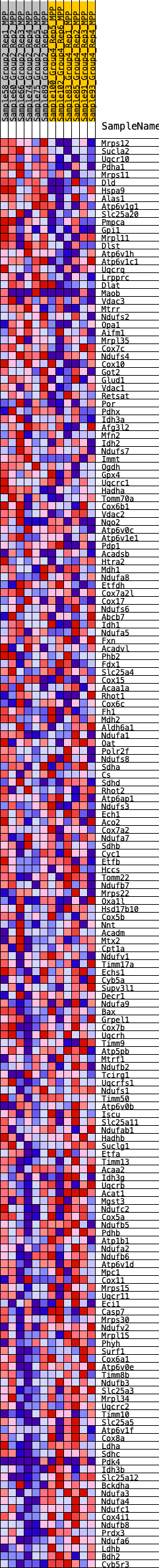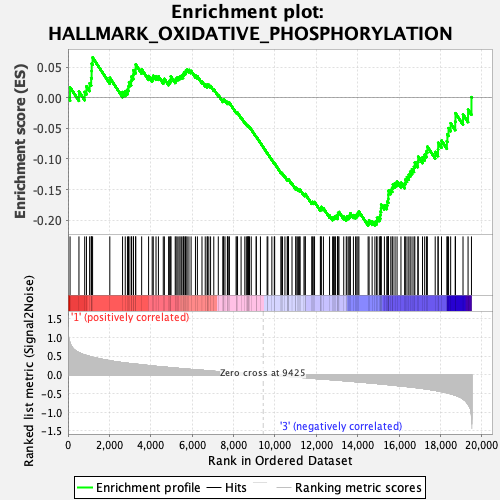
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | -0.2089618 |
| Normalized Enrichment Score (NES) | -0.763425 |
| Nominal p-value | 0.7214429 |
| FDR q-value | 0.91226274 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mrps12 | 99 | 0.845 | 0.0169 | No |
| 2 | Sucla2 | 529 | 0.586 | 0.0100 | No |
| 3 | Uqcr10 | 807 | 0.528 | 0.0095 | No |
| 4 | Pdha1 | 892 | 0.514 | 0.0186 | No |
| 5 | Mrps11 | 1045 | 0.489 | 0.0235 | No |
| 6 | Dld | 1119 | 0.479 | 0.0322 | No |
| 7 | Hspa9 | 1141 | 0.476 | 0.0435 | No |
| 8 | Alas1 | 1146 | 0.475 | 0.0557 | No |
| 9 | Atp6v1g1 | 1188 | 0.467 | 0.0658 | No |
| 10 | Slc25a20 | 2020 | 0.374 | 0.0326 | No |
| 11 | Pmpca | 2636 | 0.324 | 0.0092 | No |
| 12 | Gpi1 | 2766 | 0.314 | 0.0107 | No |
| 13 | Mrpl11 | 2873 | 0.306 | 0.0132 | No |
| 14 | Dlst | 2924 | 0.304 | 0.0186 | No |
| 15 | Atp6v1h | 2950 | 0.302 | 0.0252 | No |
| 16 | Atp6v1c1 | 3050 | 0.297 | 0.0278 | No |
| 17 | Uqcrq | 3068 | 0.296 | 0.0346 | No |
| 18 | Lrpprc | 3156 | 0.289 | 0.0377 | No |
| 19 | Dlat | 3164 | 0.289 | 0.0449 | No |
| 20 | Maob | 3269 | 0.282 | 0.0469 | No |
| 21 | Vdac3 | 3270 | 0.282 | 0.0542 | No |
| 22 | Mtrr | 3559 | 0.266 | 0.0463 | No |
| 23 | Ndufs2 | 3894 | 0.245 | 0.0354 | No |
| 24 | Opa1 | 4068 | 0.235 | 0.0326 | No |
| 25 | Aifm1 | 4120 | 0.233 | 0.0360 | No |
| 26 | Mrpl35 | 4253 | 0.224 | 0.0350 | No |
| 27 | Cox7c | 4369 | 0.219 | 0.0348 | No |
| 28 | Ndufs4 | 4603 | 0.208 | 0.0282 | No |
| 29 | Cox10 | 4657 | 0.205 | 0.0308 | No |
| 30 | Got2 | 4861 | 0.194 | 0.0254 | No |
| 31 | Glud1 | 4912 | 0.192 | 0.0278 | No |
| 32 | Vdac1 | 4967 | 0.189 | 0.0299 | No |
| 33 | Retsat | 4969 | 0.189 | 0.0348 | No |
| 34 | Por | 5181 | 0.179 | 0.0286 | No |
| 35 | Pdhx | 5220 | 0.177 | 0.0312 | No |
| 36 | Idh3a | 5275 | 0.174 | 0.0330 | No |
| 37 | Afg3l2 | 5358 | 0.169 | 0.0332 | No |
| 38 | Mfn2 | 5414 | 0.167 | 0.0347 | No |
| 39 | Idh2 | 5476 | 0.164 | 0.0358 | No |
| 40 | Ndufs7 | 5557 | 0.160 | 0.0358 | No |
| 41 | Immt | 5576 | 0.159 | 0.0390 | No |
| 42 | Ogdh | 5611 | 0.158 | 0.0414 | No |
| 43 | Gpx4 | 5676 | 0.154 | 0.0421 | No |
| 44 | Uqcrc1 | 5703 | 0.153 | 0.0448 | No |
| 45 | Hadha | 5756 | 0.151 | 0.0460 | No |
| 46 | Tomm70a | 5844 | 0.147 | 0.0454 | No |
| 47 | Cox6b1 | 5948 | 0.141 | 0.0437 | No |
| 48 | Vdac2 | 6159 | 0.132 | 0.0363 | No |
| 49 | Nqo2 | 6252 | 0.127 | 0.0348 | No |
| 50 | Atp6v0c | 6474 | 0.119 | 0.0265 | No |
| 51 | Atp6v1e1 | 6631 | 0.112 | 0.0214 | No |
| 52 | Pdp1 | 6724 | 0.107 | 0.0194 | No |
| 53 | Acadsb | 6734 | 0.107 | 0.0217 | No |
| 54 | Htra2 | 6804 | 0.103 | 0.0208 | No |
| 55 | Mdh1 | 6889 | 0.099 | 0.0191 | No |
| 56 | Ndufa8 | 7044 | 0.092 | 0.0135 | No |
| 57 | Etfdh | 7262 | 0.083 | 0.0045 | No |
| 58 | Cox7a2l | 7486 | 0.075 | -0.0051 | No |
| 59 | Cox17 | 7498 | 0.075 | -0.0037 | No |
| 60 | Ndufs6 | 7522 | 0.074 | -0.0030 | No |
| 61 | Abcb7 | 7586 | 0.072 | -0.0044 | No |
| 62 | Idh1 | 7702 | 0.067 | -0.0085 | No |
| 63 | Ndufa5 | 7706 | 0.067 | -0.0070 | No |
| 64 | Fxn | 7769 | 0.065 | -0.0085 | No |
| 65 | Acadvl | 7798 | 0.064 | -0.0082 | No |
| 66 | Phb2 | 8122 | 0.050 | -0.0236 | No |
| 67 | Fdx1 | 8162 | 0.049 | -0.0244 | No |
| 68 | Slc25a4 | 8192 | 0.047 | -0.0247 | No |
| 69 | Cox15 | 8363 | 0.041 | -0.0324 | No |
| 70 | Acaa1a | 8544 | 0.033 | -0.0408 | No |
| 71 | Rhot1 | 8616 | 0.030 | -0.0437 | No |
| 72 | Cox6c | 8665 | 0.028 | -0.0455 | No |
| 73 | Fh1 | 8689 | 0.027 | -0.0460 | No |
| 74 | Mdh2 | 8717 | 0.026 | -0.0467 | No |
| 75 | Aldh6a1 | 8739 | 0.025 | -0.0471 | No |
| 76 | Ndufa1 | 8769 | 0.024 | -0.0480 | No |
| 77 | Oat | 8854 | 0.020 | -0.0518 | No |
| 78 | Polr2f | 9094 | 0.012 | -0.0639 | No |
| 79 | Ndufs8 | 9097 | 0.012 | -0.0637 | No |
| 80 | Sdha | 9299 | 0.005 | -0.0739 | No |
| 81 | Cs | 9621 | -0.006 | -0.0904 | No |
| 82 | Sdhd | 9656 | -0.007 | -0.0920 | No |
| 83 | Rhot2 | 9851 | -0.014 | -0.1017 | No |
| 84 | Atp6ap1 | 9968 | -0.019 | -0.1071 | No |
| 85 | Ndufs3 | 9986 | -0.020 | -0.1075 | No |
| 86 | Ech1 | 10290 | -0.032 | -0.1223 | No |
| 87 | Aco2 | 10304 | -0.033 | -0.1222 | No |
| 88 | Cox7a2 | 10366 | -0.035 | -0.1244 | No |
| 89 | Ndufa7 | 10482 | -0.040 | -0.1293 | No |
| 90 | Sdhb | 10591 | -0.044 | -0.1337 | No |
| 91 | Cyc1 | 10613 | -0.044 | -0.1337 | No |
| 92 | Etfb | 10647 | -0.046 | -0.1342 | No |
| 93 | Hccs | 10654 | -0.046 | -0.1333 | No |
| 94 | Tomm22 | 10824 | -0.053 | -0.1407 | No |
| 95 | Ndufb7 | 11007 | -0.060 | -0.1485 | No |
| 96 | Mrps22 | 11014 | -0.060 | -0.1473 | No |
| 97 | Oxa1l | 11080 | -0.063 | -0.1490 | No |
| 98 | Hsd17b10 | 11133 | -0.065 | -0.1500 | No |
| 99 | Cox5b | 11193 | -0.068 | -0.1512 | No |
| 100 | Nnt | 11215 | -0.068 | -0.1506 | No |
| 101 | Acadm | 11402 | -0.075 | -0.1582 | No |
| 102 | Mtx2 | 11456 | -0.077 | -0.1589 | No |
| 103 | Cpt1a | 11465 | -0.078 | -0.1573 | No |
| 104 | Ndufv1 | 11778 | -0.092 | -0.1710 | No |
| 105 | Timm17a | 11809 | -0.094 | -0.1702 | No |
| 106 | Echs1 | 11875 | -0.096 | -0.1710 | No |
| 107 | Cyb5a | 11913 | -0.098 | -0.1704 | No |
| 108 | Supv3l1 | 12194 | -0.110 | -0.1820 | No |
| 109 | Decr1 | 12221 | -0.111 | -0.1804 | No |
| 110 | Ndufa9 | 12239 | -0.112 | -0.1784 | No |
| 111 | Bax | 12342 | -0.116 | -0.1806 | No |
| 112 | Grpel1 | 12638 | -0.126 | -0.1926 | No |
| 113 | Cox7b | 12786 | -0.132 | -0.1968 | No |
| 114 | Uqcrh | 12826 | -0.133 | -0.1953 | No |
| 115 | Timm9 | 12884 | -0.136 | -0.1947 | No |
| 116 | Atp5pb | 12929 | -0.138 | -0.1934 | No |
| 117 | Mtrf1 | 13022 | -0.142 | -0.1944 | No |
| 118 | Ndufb2 | 13033 | -0.143 | -0.1912 | No |
| 119 | Tcirg1 | 13053 | -0.144 | -0.1884 | No |
| 120 | Uqcrfs1 | 13094 | -0.145 | -0.1867 | No |
| 121 | Ndufs1 | 13323 | -0.157 | -0.1944 | No |
| 122 | Timm50 | 13445 | -0.163 | -0.1964 | No |
| 123 | Atp6v0b | 13481 | -0.164 | -0.1940 | No |
| 124 | Iscu | 13566 | -0.167 | -0.1939 | No |
| 125 | Slc25a11 | 13617 | -0.170 | -0.1921 | No |
| 126 | Ndufab1 | 13643 | -0.171 | -0.1889 | No |
| 127 | Hadhb | 13791 | -0.177 | -0.1919 | No |
| 128 | Suclg1 | 13896 | -0.182 | -0.1925 | No |
| 129 | Etfa | 13956 | -0.185 | -0.1907 | No |
| 130 | Timm13 | 14010 | -0.187 | -0.1886 | No |
| 131 | Acaa2 | 14057 | -0.190 | -0.1860 | No |
| 132 | Idh3g | 14502 | -0.211 | -0.2034 | Yes |
| 133 | Uqcrb | 14549 | -0.214 | -0.2002 | Yes |
| 134 | Acat1 | 14696 | -0.220 | -0.2020 | Yes |
| 135 | Mgst3 | 14829 | -0.227 | -0.2029 | Yes |
| 136 | Ndufc2 | 14913 | -0.232 | -0.2012 | Yes |
| 137 | Cox5a | 14933 | -0.233 | -0.1961 | Yes |
| 138 | Ndufb5 | 15043 | -0.239 | -0.1955 | Yes |
| 139 | Pdhb | 15089 | -0.242 | -0.1915 | Yes |
| 140 | Atp1b1 | 15103 | -0.242 | -0.1859 | Yes |
| 141 | Ndufa2 | 15126 | -0.243 | -0.1806 | Yes |
| 142 | Ndufb6 | 15132 | -0.243 | -0.1746 | Yes |
| 143 | Atp6v1d | 15288 | -0.252 | -0.1760 | Yes |
| 144 | Mpc1 | 15407 | -0.258 | -0.1754 | Yes |
| 145 | Cox11 | 15428 | -0.259 | -0.1696 | Yes |
| 146 | Mrps15 | 15474 | -0.262 | -0.1651 | Yes |
| 147 | Uqcr11 | 15479 | -0.262 | -0.1585 | Yes |
| 148 | Eci1 | 15483 | -0.262 | -0.1518 | Yes |
| 149 | Casp7 | 15597 | -0.269 | -0.1507 | Yes |
| 150 | Mrps30 | 15665 | -0.272 | -0.1470 | Yes |
| 151 | Ndufv2 | 15701 | -0.275 | -0.1416 | Yes |
| 152 | Mrpl15 | 15803 | -0.281 | -0.1395 | Yes |
| 153 | Phyh | 15900 | -0.287 | -0.1370 | Yes |
| 154 | Surf1 | 16088 | -0.298 | -0.1389 | Yes |
| 155 | Cox6a1 | 16267 | -0.306 | -0.1401 | Yes |
| 156 | Atp6v0e | 16299 | -0.309 | -0.1337 | Yes |
| 157 | Timm8b | 16380 | -0.314 | -0.1296 | Yes |
| 158 | Ndufb3 | 16468 | -0.318 | -0.1258 | Yes |
| 159 | Slc25a3 | 16535 | -0.322 | -0.1208 | Yes |
| 160 | Mrpl34 | 16623 | -0.328 | -0.1167 | Yes |
| 161 | Uqcrc2 | 16718 | -0.335 | -0.1128 | Yes |
| 162 | Timm10 | 16758 | -0.337 | -0.1061 | Yes |
| 163 | Slc25a5 | 16904 | -0.349 | -0.1045 | Yes |
| 164 | Atp6v1f | 16922 | -0.350 | -0.0962 | Yes |
| 165 | Cox8a | 17130 | -0.363 | -0.0974 | Yes |
| 166 | Ldha | 17234 | -0.371 | -0.0931 | Yes |
| 167 | Sdhc | 17320 | -0.377 | -0.0876 | Yes |
| 168 | Pdk4 | 17364 | -0.381 | -0.0799 | Yes |
| 169 | Idh3b | 17743 | -0.415 | -0.0886 | Yes |
| 170 | Slc25a12 | 17879 | -0.429 | -0.0844 | Yes |
| 171 | Bckdha | 17887 | -0.430 | -0.0735 | Yes |
| 172 | Ndufa3 | 18047 | -0.447 | -0.0701 | Yes |
| 173 | Ndufa4 | 18308 | -0.478 | -0.0710 | Yes |
| 174 | Ndufc1 | 18333 | -0.481 | -0.0597 | Yes |
| 175 | Cox4i1 | 18388 | -0.490 | -0.0497 | Yes |
| 176 | Ndufb8 | 18490 | -0.506 | -0.0418 | Yes |
| 177 | Prdx3 | 18710 | -0.542 | -0.0389 | Yes |
| 178 | Ndufa6 | 18723 | -0.545 | -0.0253 | Yes |
| 179 | Ldhb | 19087 | -0.641 | -0.0274 | Yes |
| 180 | Bdh2 | 19331 | -0.787 | -0.0194 | Yes |
| 181 | Cyb5r3 | 19495 | -1.100 | 0.0009 | Yes |