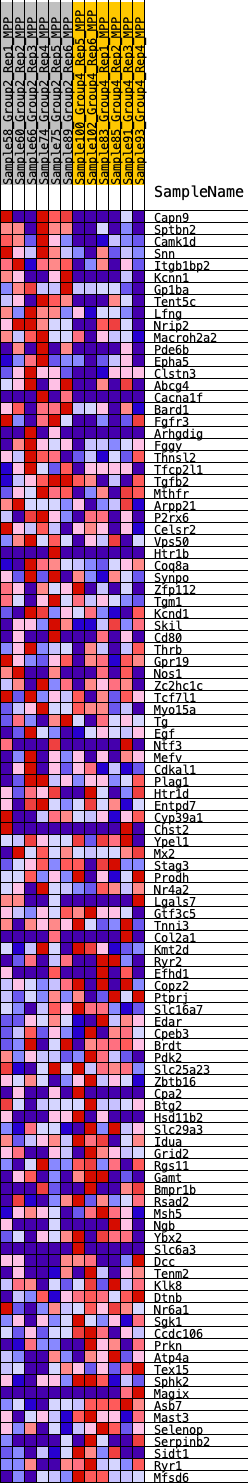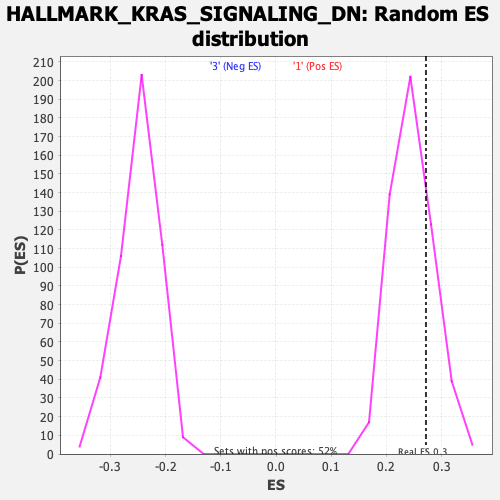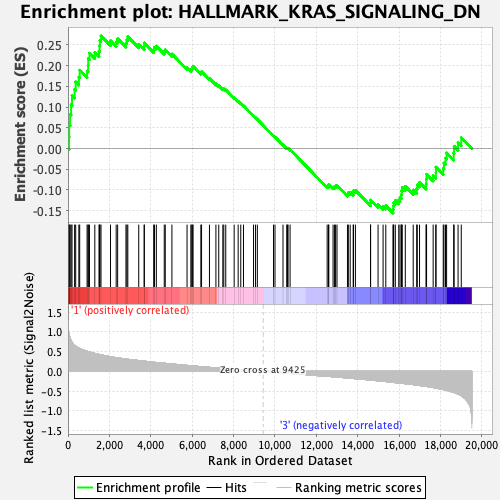
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.2716799 |
| Normalized Enrichment Score (NES) | 1.1074024 |
| Nominal p-value | 0.26285714 |
| FDR q-value | 0.9155124 |
| FWER p-Value | 0.985 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Capn9 | 52 | 0.934 | 0.0278 | Yes |
| 2 | Sptbn2 | 58 | 0.922 | 0.0576 | Yes |
| 3 | Camk1d | 103 | 0.842 | 0.0827 | Yes |
| 4 | Snn | 149 | 0.778 | 0.1058 | Yes |
| 5 | Itgb1bp2 | 194 | 0.737 | 0.1275 | Yes |
| 6 | Kcnn1 | 319 | 0.660 | 0.1426 | Yes |
| 7 | Gp1ba | 372 | 0.639 | 0.1607 | Yes |
| 8 | Tent5c | 525 | 0.588 | 0.1721 | Yes |
| 9 | Lfng | 565 | 0.579 | 0.1890 | Yes |
| 10 | Nrip2 | 922 | 0.508 | 0.1872 | Yes |
| 11 | Macroh2a2 | 973 | 0.502 | 0.2009 | Yes |
| 12 | Pde6b | 978 | 0.500 | 0.2170 | Yes |
| 13 | Epha5 | 1037 | 0.491 | 0.2300 | Yes |
| 14 | Clstn3 | 1296 | 0.455 | 0.2316 | Yes |
| 15 | Abcg4 | 1498 | 0.429 | 0.2352 | Yes |
| 16 | Cacna1f | 1528 | 0.426 | 0.2476 | Yes |
| 17 | Bard1 | 1542 | 0.424 | 0.2607 | Yes |
| 18 | Fgfr3 | 1594 | 0.418 | 0.2717 | Yes |
| 19 | Arhgdig | 2053 | 0.371 | 0.2602 | No |
| 20 | Fggy | 2328 | 0.346 | 0.2573 | No |
| 21 | Thnsl2 | 2399 | 0.341 | 0.2648 | No |
| 22 | Tfcp2l1 | 2800 | 0.311 | 0.2544 | No |
| 23 | Tgfb2 | 2846 | 0.308 | 0.2621 | No |
| 24 | Mthfr | 2890 | 0.306 | 0.2698 | No |
| 25 | Arpp21 | 3418 | 0.276 | 0.2517 | No |
| 26 | P2rx6 | 3682 | 0.258 | 0.2465 | No |
| 27 | Celsr2 | 3686 | 0.258 | 0.2547 | No |
| 28 | Vps50 | 4141 | 0.231 | 0.2389 | No |
| 29 | Htr1b | 4179 | 0.229 | 0.2444 | No |
| 30 | Coq8a | 4268 | 0.223 | 0.2472 | No |
| 31 | Synpo | 4651 | 0.206 | 0.2342 | No |
| 32 | Zfp112 | 4699 | 0.203 | 0.2384 | No |
| 33 | Tgm1 | 5021 | 0.187 | 0.2279 | No |
| 34 | Kcnd1 | 5752 | 0.151 | 0.1952 | No |
| 35 | Skil | 5936 | 0.141 | 0.1904 | No |
| 36 | Cd80 | 5981 | 0.139 | 0.1927 | No |
| 37 | Thrb | 6009 | 0.138 | 0.1958 | No |
| 38 | Gpr19 | 6044 | 0.136 | 0.1985 | No |
| 39 | Nos1 | 6420 | 0.121 | 0.1831 | No |
| 40 | Zc2hc1c | 6453 | 0.120 | 0.1854 | No |
| 41 | Tcf7l1 | 6835 | 0.101 | 0.1690 | No |
| 42 | Myo15a | 7147 | 0.088 | 0.1559 | No |
| 43 | Tg | 7284 | 0.082 | 0.1515 | No |
| 44 | Egf | 7481 | 0.076 | 0.1439 | No |
| 45 | Ntf3 | 7528 | 0.074 | 0.1439 | No |
| 46 | Mefv | 7623 | 0.070 | 0.1414 | No |
| 47 | Cdkal1 | 8029 | 0.054 | 0.1223 | No |
| 48 | Plag1 | 8222 | 0.046 | 0.1139 | No |
| 49 | Htr1d | 8348 | 0.042 | 0.1088 | No |
| 50 | Entpd7 | 8479 | 0.036 | 0.1033 | No |
| 51 | Cyp39a1 | 8965 | 0.016 | 0.0788 | No |
| 52 | Chst2 | 9062 | 0.013 | 0.0743 | No |
| 53 | Ypel1 | 9149 | 0.010 | 0.0702 | No |
| 54 | Mx2 | 9930 | -0.018 | 0.0306 | No |
| 55 | Stag3 | 10002 | -0.021 | 0.0276 | No |
| 56 | Prodh | 10389 | -0.037 | 0.0089 | No |
| 57 | Nr4a2 | 10569 | -0.043 | 0.0011 | No |
| 58 | Lgals7 | 10604 | -0.044 | 0.0007 | No |
| 59 | Gtf3c5 | 10634 | -0.045 | 0.0007 | No |
| 60 | Tnni3 | 10731 | -0.048 | -0.0026 | No |
| 61 | Col2a1 | 12523 | -0.123 | -0.0909 | No |
| 62 | Kmt2d | 12587 | -0.124 | -0.0901 | No |
| 63 | Ryr2 | 12603 | -0.125 | -0.0868 | No |
| 64 | Efhd1 | 12808 | -0.133 | -0.0930 | No |
| 65 | Copz2 | 12880 | -0.136 | -0.0922 | No |
| 66 | Ptprj | 12930 | -0.138 | -0.0903 | No |
| 67 | Slc16a7 | 12998 | -0.141 | -0.0891 | No |
| 68 | Edar | 13504 | -0.165 | -0.1098 | No |
| 69 | Cpeb3 | 13545 | -0.167 | -0.1064 | No |
| 70 | Brdt | 13635 | -0.171 | -0.1054 | No |
| 71 | Pdk2 | 13780 | -0.177 | -0.1071 | No |
| 72 | Slc25a23 | 13795 | -0.177 | -0.1020 | No |
| 73 | Zbtb16 | 13887 | -0.181 | -0.1008 | No |
| 74 | Cpa2 | 14618 | -0.217 | -0.1313 | No |
| 75 | Btg2 | 14622 | -0.217 | -0.1244 | No |
| 76 | Hsd11b2 | 14982 | -0.236 | -0.1352 | No |
| 77 | Slc29a3 | 15225 | -0.249 | -0.1396 | No |
| 78 | Idua | 15356 | -0.255 | -0.1380 | No |
| 79 | Grid2 | 15706 | -0.275 | -0.1470 | No |
| 80 | Rgs11 | 15721 | -0.276 | -0.1387 | No |
| 81 | Gamt | 15738 | -0.277 | -0.1305 | No |
| 82 | Bmpr1b | 15816 | -0.282 | -0.1253 | No |
| 83 | Rsad2 | 15982 | -0.292 | -0.1243 | No |
| 84 | Msh5 | 16043 | -0.295 | -0.1177 | No |
| 85 | Ngb | 16109 | -0.299 | -0.1113 | No |
| 86 | Ybx2 | 16120 | -0.300 | -0.1021 | No |
| 87 | Slc6a3 | 16150 | -0.301 | -0.0938 | No |
| 88 | Dcc | 16298 | -0.309 | -0.0913 | No |
| 89 | Tenm2 | 16680 | -0.332 | -0.1001 | No |
| 90 | Klk8 | 16849 | -0.344 | -0.0976 | No |
| 91 | Dtnb | 16876 | -0.347 | -0.0876 | No |
| 92 | Nr6a1 | 16986 | -0.354 | -0.0817 | No |
| 93 | Sgk1 | 17305 | -0.376 | -0.0858 | No |
| 94 | Ccdc106 | 17312 | -0.377 | -0.0738 | No |
| 95 | Prkn | 17326 | -0.378 | -0.0622 | No |
| 96 | Atp4a | 17644 | -0.407 | -0.0653 | No |
| 97 | Tex15 | 17776 | -0.418 | -0.0584 | No |
| 98 | Sphk2 | 17778 | -0.418 | -0.0448 | No |
| 99 | Magix | 18131 | -0.456 | -0.0481 | No |
| 100 | Asb7 | 18165 | -0.459 | -0.0349 | No |
| 101 | Mast3 | 18241 | -0.470 | -0.0234 | No |
| 102 | Selenop | 18294 | -0.476 | -0.0106 | No |
| 103 | Serpinb2 | 18638 | -0.530 | -0.0110 | No |
| 104 | Sidt1 | 18655 | -0.533 | 0.0056 | No |
| 105 | Ryr1 | 18848 | -0.571 | 0.0143 | No |
| 106 | Mfsd6 | 19002 | -0.610 | 0.0263 | No |