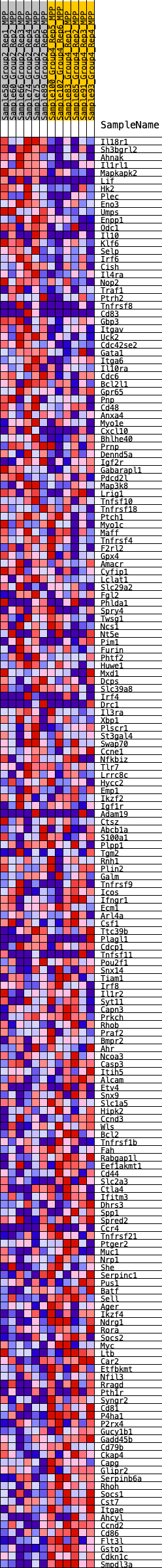
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
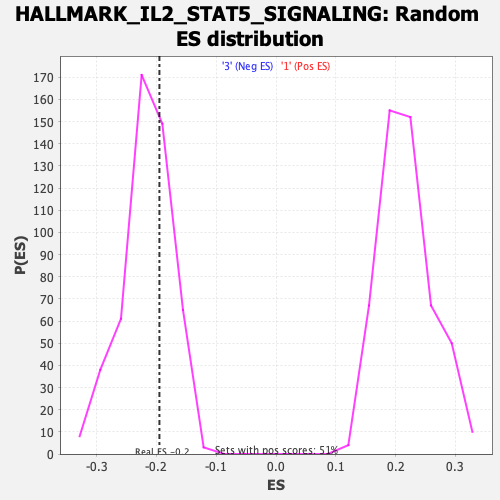
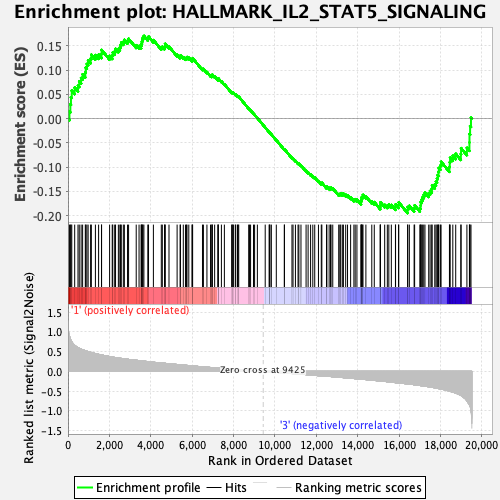
| GeneSet | HALLMARK_IL2_STAT5_SIGNALING |
| Enrichment Score (ES) | -0.19508262 |
| Normalized Enrichment Score (NES) | -0.9049843 |
| Nominal p-value | 0.66868687 |
| FDR q-value | 0.8875655 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Il18r1 | 70 | 0.896 | 0.0148 | No |
| 2 | Sh3bgrl2 | 114 | 0.826 | 0.0295 | No |
| 3 | Ahnak | 143 | 0.785 | 0.0442 | No |
| 4 | Il1rl1 | 182 | 0.750 | 0.0576 | No |
| 5 | Mapkapk2 | 323 | 0.658 | 0.0639 | No |
| 6 | Lif | 483 | 0.601 | 0.0680 | No |
| 7 | Hk2 | 543 | 0.583 | 0.0769 | No |
| 8 | Plec | 634 | 0.564 | 0.0839 | No |
| 9 | Eno3 | 706 | 0.546 | 0.0914 | No |
| 10 | Umps | 836 | 0.525 | 0.0955 | No |
| 11 | Enpp1 | 858 | 0.521 | 0.1051 | No |
| 12 | Odc1 | 912 | 0.510 | 0.1128 | No |
| 13 | Il10 | 972 | 0.502 | 0.1201 | No |
| 14 | Klf6 | 1093 | 0.483 | 0.1238 | No |
| 15 | Selp | 1127 | 0.477 | 0.1319 | No |
| 16 | Irf6 | 1324 | 0.451 | 0.1310 | No |
| 17 | Cish | 1479 | 0.431 | 0.1319 | No |
| 18 | Il4ra | 1618 | 0.415 | 0.1333 | No |
| 19 | Nop2 | 1623 | 0.414 | 0.1416 | No |
| 20 | Traf1 | 2010 | 0.375 | 0.1293 | No |
| 21 | Ptrh2 | 2137 | 0.362 | 0.1303 | No |
| 22 | Tnfrsf8 | 2163 | 0.359 | 0.1364 | No |
| 23 | Cd83 | 2257 | 0.352 | 0.1388 | No |
| 24 | Gbp3 | 2297 | 0.348 | 0.1439 | No |
| 25 | Itgav | 2434 | 0.338 | 0.1438 | No |
| 26 | Uck2 | 2506 | 0.333 | 0.1470 | No |
| 27 | Cdc42se2 | 2540 | 0.330 | 0.1520 | No |
| 28 | Gata1 | 2574 | 0.328 | 0.1571 | No |
| 29 | Itga6 | 2673 | 0.320 | 0.1586 | No |
| 30 | Il10ra | 2723 | 0.317 | 0.1625 | No |
| 31 | Cdc6 | 2877 | 0.306 | 0.1609 | No |
| 32 | Bcl2l1 | 2927 | 0.303 | 0.1646 | No |
| 33 | Gpr65 | 3296 | 0.281 | 0.1513 | No |
| 34 | Pnp | 3433 | 0.275 | 0.1499 | No |
| 35 | Cd48 | 3533 | 0.268 | 0.1503 | No |
| 36 | Anxa4 | 3549 | 0.266 | 0.1550 | No |
| 37 | Myo1e | 3573 | 0.265 | 0.1593 | No |
| 38 | Cxcl10 | 3592 | 0.263 | 0.1637 | No |
| 39 | Bhlhe40 | 3621 | 0.262 | 0.1677 | No |
| 40 | Prnp | 3667 | 0.259 | 0.1707 | No |
| 41 | Dennd5a | 3857 | 0.247 | 0.1659 | No |
| 42 | Igf2r | 3891 | 0.245 | 0.1693 | No |
| 43 | Gabarapl1 | 4126 | 0.232 | 0.1619 | No |
| 44 | Pdcd2l | 4508 | 0.213 | 0.1466 | No |
| 45 | Map3k8 | 4557 | 0.211 | 0.1484 | No |
| 46 | Lrig1 | 4664 | 0.205 | 0.1472 | No |
| 47 | Tnfsf10 | 4689 | 0.204 | 0.1501 | No |
| 48 | Tnfrsf18 | 4691 | 0.204 | 0.1542 | No |
| 49 | Ptch1 | 4880 | 0.194 | 0.1485 | No |
| 50 | Myo1c | 5272 | 0.174 | 0.1318 | No |
| 51 | Maff | 5416 | 0.167 | 0.1279 | No |
| 52 | Tnfrsf4 | 5435 | 0.166 | 0.1303 | No |
| 53 | F2rl2 | 5571 | 0.159 | 0.1266 | No |
| 54 | Gpx4 | 5676 | 0.154 | 0.1244 | No |
| 55 | Amacr | 5729 | 0.152 | 0.1249 | No |
| 56 | Cyfip1 | 5748 | 0.151 | 0.1270 | No |
| 57 | Lclat1 | 5837 | 0.147 | 0.1255 | No |
| 58 | Slc29a2 | 5996 | 0.138 | 0.1202 | No |
| 59 | Fgl2 | 6004 | 0.138 | 0.1226 | No |
| 60 | Phlda1 | 6022 | 0.138 | 0.1246 | No |
| 61 | Spry4 | 6498 | 0.118 | 0.1024 | No |
| 62 | Twsg1 | 6543 | 0.116 | 0.1025 | No |
| 63 | Ncs1 | 6713 | 0.108 | 0.0960 | No |
| 64 | Nt5e | 6887 | 0.099 | 0.0891 | No |
| 65 | Pim1 | 6954 | 0.096 | 0.0876 | No |
| 66 | Furin | 6959 | 0.096 | 0.0894 | No |
| 67 | Phtf2 | 6971 | 0.095 | 0.0908 | No |
| 68 | Huwe1 | 7086 | 0.090 | 0.0868 | No |
| 69 | Mxd1 | 7251 | 0.084 | 0.0800 | No |
| 70 | Dcps | 7252 | 0.083 | 0.0817 | No |
| 71 | Slc39a8 | 7260 | 0.083 | 0.0830 | No |
| 72 | Irf4 | 7412 | 0.078 | 0.0768 | No |
| 73 | Drc1 | 7556 | 0.073 | 0.0709 | No |
| 74 | Il3ra | 7909 | 0.059 | 0.0539 | No |
| 75 | Xbp1 | 7932 | 0.058 | 0.0540 | No |
| 76 | Plscr1 | 7964 | 0.056 | 0.0535 | No |
| 77 | St3gal4 | 7991 | 0.055 | 0.0533 | No |
| 78 | Swap70 | 8083 | 0.052 | 0.0497 | No |
| 79 | Ccne1 | 8105 | 0.051 | 0.0496 | No |
| 80 | Nfkbiz | 8190 | 0.047 | 0.0462 | No |
| 81 | Tlr7 | 8218 | 0.046 | 0.0458 | No |
| 82 | Lrrc8c | 8249 | 0.045 | 0.0452 | No |
| 83 | Hycc2 | 8731 | 0.025 | 0.0208 | No |
| 84 | Emp1 | 8772 | 0.023 | 0.0192 | No |
| 85 | Ikzf2 | 8822 | 0.021 | 0.0171 | No |
| 86 | Igf1r | 8964 | 0.016 | 0.0102 | No |
| 87 | Adam19 | 9016 | 0.014 | 0.0078 | No |
| 88 | Ctsz | 9150 | 0.010 | 0.0011 | No |
| 89 | Abcb1a | 9529 | -0.001 | -0.0184 | No |
| 90 | S100a1 | 9718 | -0.009 | -0.0279 | No |
| 91 | Plpp1 | 9744 | -0.010 | -0.0290 | No |
| 92 | Tgm2 | 9755 | -0.010 | -0.0293 | No |
| 93 | Rnh1 | 9826 | -0.013 | -0.0327 | No |
| 94 | Plin2 | 10065 | -0.023 | -0.0445 | No |
| 95 | Galm | 10452 | -0.039 | -0.0637 | No |
| 96 | Tnfrsf9 | 10459 | -0.039 | -0.0632 | No |
| 97 | Icos | 10822 | -0.053 | -0.0808 | No |
| 98 | Ifngr1 | 10873 | -0.055 | -0.0823 | No |
| 99 | Ecm1 | 10989 | -0.059 | -0.0870 | No |
| 100 | Arl4a | 11116 | -0.064 | -0.0922 | No |
| 101 | Csf1 | 11155 | -0.066 | -0.0928 | No |
| 102 | Ttc39b | 11257 | -0.070 | -0.0966 | No |
| 103 | Plagl1 | 11504 | -0.079 | -0.1077 | No |
| 104 | Cdcp1 | 11602 | -0.084 | -0.1110 | No |
| 105 | Tnfsf11 | 11722 | -0.089 | -0.1153 | No |
| 106 | Pou2f1 | 11829 | -0.095 | -0.1189 | No |
| 107 | Snx14 | 11922 | -0.098 | -0.1216 | No |
| 108 | Tiam1 | 12102 | -0.106 | -0.1287 | No |
| 109 | Irf8 | 12238 | -0.112 | -0.1334 | No |
| 110 | Il1r2 | 12266 | -0.113 | -0.1325 | No |
| 111 | Syt11 | 12490 | -0.122 | -0.1415 | No |
| 112 | Capn3 | 12509 | -0.122 | -0.1399 | No |
| 113 | Prkch | 12618 | -0.125 | -0.1429 | No |
| 114 | Rhob | 12685 | -0.128 | -0.1437 | No |
| 115 | Praf2 | 12710 | -0.129 | -0.1423 | No |
| 116 | Bmpr2 | 12793 | -0.132 | -0.1438 | No |
| 117 | Ahr | 13081 | -0.145 | -0.1557 | No |
| 118 | Ncoa3 | 13135 | -0.147 | -0.1555 | No |
| 119 | Casp3 | 13176 | -0.149 | -0.1545 | No |
| 120 | Itih5 | 13255 | -0.153 | -0.1554 | No |
| 121 | Alcam | 13307 | -0.155 | -0.1548 | No |
| 122 | Etv4 | 13418 | -0.161 | -0.1572 | No |
| 123 | Snx9 | 13507 | -0.165 | -0.1583 | No |
| 124 | Slc1a5 | 13656 | -0.172 | -0.1625 | No |
| 125 | Hipk2 | 13815 | -0.179 | -0.1670 | No |
| 126 | Ccnd3 | 13876 | -0.181 | -0.1664 | No |
| 127 | Wls | 13961 | -0.185 | -0.1669 | No |
| 128 | Bcl2 | 14158 | -0.194 | -0.1731 | No |
| 129 | Tnfrsf1b | 14162 | -0.195 | -0.1692 | No |
| 130 | Fah | 14182 | -0.195 | -0.1662 | No |
| 131 | Rabgap1l | 14188 | -0.196 | -0.1624 | No |
| 132 | Eef1akmt1 | 14237 | -0.198 | -0.1608 | No |
| 133 | Cd44 | 14250 | -0.199 | -0.1574 | No |
| 134 | Slc2a3 | 14394 | -0.206 | -0.1605 | No |
| 135 | Ctla4 | 14679 | -0.220 | -0.1707 | No |
| 136 | Ifitm3 | 14799 | -0.225 | -0.1723 | No |
| 137 | Dhrs3 | 15086 | -0.242 | -0.1821 | No |
| 138 | Spp1 | 15092 | -0.242 | -0.1774 | No |
| 139 | Spred2 | 15098 | -0.242 | -0.1727 | No |
| 140 | Ccr4 | 15302 | -0.252 | -0.1780 | No |
| 141 | Tnfrsf21 | 15430 | -0.259 | -0.1792 | No |
| 142 | Ptger2 | 15502 | -0.263 | -0.1775 | No |
| 143 | Muc1 | 15623 | -0.270 | -0.1782 | No |
| 144 | Nrp1 | 15822 | -0.283 | -0.1826 | No |
| 145 | She | 15828 | -0.283 | -0.1771 | No |
| 146 | Serpinc1 | 15973 | -0.292 | -0.1785 | No |
| 147 | Pus1 | 15981 | -0.292 | -0.1729 | No |
| 148 | Batf | 16411 | -0.316 | -0.1886 | Yes |
| 149 | Sell | 16416 | -0.316 | -0.1823 | Yes |
| 150 | Ager | 16494 | -0.320 | -0.1797 | Yes |
| 151 | Ikzf4 | 16724 | -0.335 | -0.1847 | Yes |
| 152 | Ndrg1 | 16749 | -0.336 | -0.1790 | Yes |
| 153 | Rora | 17001 | -0.355 | -0.1847 | Yes |
| 154 | Socs2 | 17039 | -0.357 | -0.1793 | Yes |
| 155 | Myc | 17043 | -0.358 | -0.1721 | Yes |
| 156 | Ltb | 17091 | -0.361 | -0.1671 | Yes |
| 157 | Car2 | 17138 | -0.364 | -0.1621 | Yes |
| 158 | Etfbkmt | 17191 | -0.368 | -0.1572 | Yes |
| 159 | Nfil3 | 17248 | -0.372 | -0.1524 | Yes |
| 160 | Rragd | 17430 | -0.386 | -0.1539 | Yes |
| 161 | Pth1r | 17496 | -0.392 | -0.1492 | Yes |
| 162 | Syngr2 | 17577 | -0.400 | -0.1451 | Yes |
| 163 | Cd81 | 17592 | -0.402 | -0.1376 | Yes |
| 164 | P4ha1 | 17723 | -0.413 | -0.1358 | Yes |
| 165 | P2rx4 | 17780 | -0.418 | -0.1301 | Yes |
| 166 | Gucy1b1 | 17830 | -0.425 | -0.1239 | Yes |
| 167 | Gadd45b | 17861 | -0.428 | -0.1167 | Yes |
| 168 | Cd79b | 17903 | -0.432 | -0.1099 | Yes |
| 169 | Ckap4 | 17918 | -0.433 | -0.1018 | Yes |
| 170 | Capg | 17995 | -0.442 | -0.0966 | Yes |
| 171 | Glipr2 | 18026 | -0.445 | -0.0890 | Yes |
| 172 | Serpinb6a | 18431 | -0.498 | -0.0997 | Yes |
| 173 | Rhoh | 18446 | -0.500 | -0.0902 | Yes |
| 174 | Socs1 | 18462 | -0.502 | -0.0806 | Yes |
| 175 | Cst7 | 18594 | -0.523 | -0.0767 | Yes |
| 176 | Itgae | 18734 | -0.547 | -0.0726 | Yes |
| 177 | Ahcyl | 18982 | -0.605 | -0.0730 | Yes |
| 178 | Ccnd2 | 18993 | -0.609 | -0.0610 | Yes |
| 179 | Cd86 | 19275 | -0.741 | -0.0603 | Yes |
| 180 | Flt3l | 19397 | -0.846 | -0.0492 | Yes |
| 181 | Gsto1 | 19400 | -0.848 | -0.0319 | Yes |
| 182 | Cdkn1c | 19430 | -0.879 | -0.0154 | Yes |
| 183 | Smpdl3a | 19472 | -0.952 | 0.0021 | Yes |