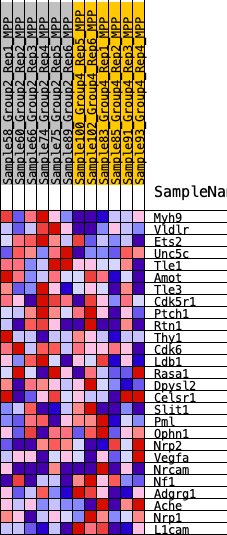
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
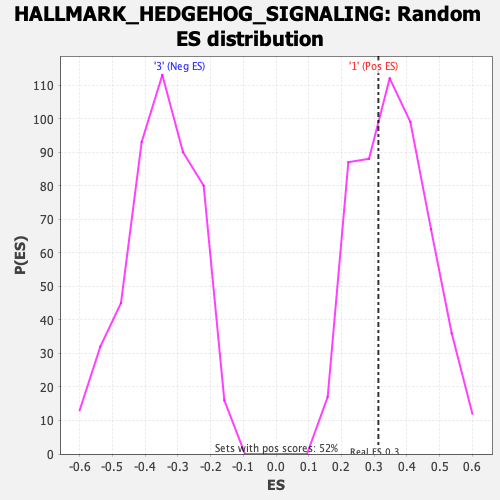
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.31291306 |
| Normalized Enrichment Score (NES) | 0.8776822 |
| Nominal p-value | 0.6351351 |
| FDR q-value | 0.9831421 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Myh9 | 394 | 0.630 | 0.0789 | Yes |
| 2 | Vldlr | 462 | 0.607 | 0.1709 | Yes |
| 3 | Ets2 | 678 | 0.551 | 0.2465 | Yes |
| 4 | Unc5c | 1665 | 0.410 | 0.2603 | Yes |
| 5 | Tle1 | 1845 | 0.393 | 0.3129 | Yes |
| 6 | Amot | 3676 | 0.258 | 0.2596 | No |
| 7 | Tle3 | 3893 | 0.245 | 0.2870 | No |
| 8 | Cdk5r1 | 4760 | 0.200 | 0.2739 | No |
| 9 | Ptch1 | 4880 | 0.194 | 0.2983 | No |
| 10 | Rtn1 | 5715 | 0.153 | 0.2795 | No |
| 11 | Thy1 | 6023 | 0.137 | 0.2853 | No |
| 12 | Cdk6 | 6833 | 0.102 | 0.2598 | No |
| 13 | Ldb1 | 6913 | 0.098 | 0.2711 | No |
| 14 | Rasa1 | 6947 | 0.096 | 0.2846 | No |
| 15 | Dpysl2 | 7970 | 0.056 | 0.2409 | No |
| 16 | Celsr1 | 9990 | -0.020 | 0.1404 | No |
| 17 | Slit1 | 11509 | -0.080 | 0.0750 | No |
| 18 | Pml | 11852 | -0.096 | 0.0725 | No |
| 19 | Ophn1 | 12285 | -0.113 | 0.0682 | No |
| 20 | Nrp2 | 12704 | -0.129 | 0.0669 | No |
| 21 | Vegfa | 13223 | -0.151 | 0.0641 | No |
| 22 | Nrcam | 13991 | -0.186 | 0.0541 | No |
| 23 | Nf1 | 14292 | -0.201 | 0.0702 | No |
| 24 | Adgrg1 | 14405 | -0.206 | 0.0969 | No |
| 25 | Ache | 15206 | -0.247 | 0.0947 | No |
| 26 | Nrp1 | 15822 | -0.283 | 0.1076 | No |
| 27 | L1cam | 18573 | -0.520 | 0.0482 | No |